- Title
-
Zebrafish Nkd1 promotes Dvl degradation and is required for left-right patterning
- Authors
- Schneider, I., Schneider, P.N., Derry, S.W., Lin, S., Barton, L.J., Westfall, T., and Slusarski, D.C.
- Source
- Full text @ Dev. Biol.
|
Expression pattern of zebrafish Nkd homologs. nkd1 expression is not detectable at 1k-cell stage (A) but ubiquitous during epiboly (B). During somite stages, nkd1 is expressed in somites, rhombomeres and tailbud at 4-somite stage (C) and in cells around the forming KV (D). nkd1 expression at 24 hpf (E). nkd2a is ubiquitously expressed during 1k-cell stage (F), epiboly (G), and in somites, rhombomeres and tailbud at 4-somite stage (H). nkd2a expression in tailbud (I) and at 24 hpf (J). nkd3 ubiquitously expressed during 1k-cell stage (K), epiboly (L), and in somites (M). nkd3 expression in tailbud (N) and at 24 hpf (O). (P–S) nkd1 is expressed ubiquitously throughout gastrulation (P) and in the FCs at 80% epiboly (Q). At 1-somite stage, marked nkd1 expression is observed the tailbud (R), and in cells around the KV (S). Arrows denote DFCs during 80% epiboly, tailbud and 5-somite stage. hpf = hours post fertilization. |
|
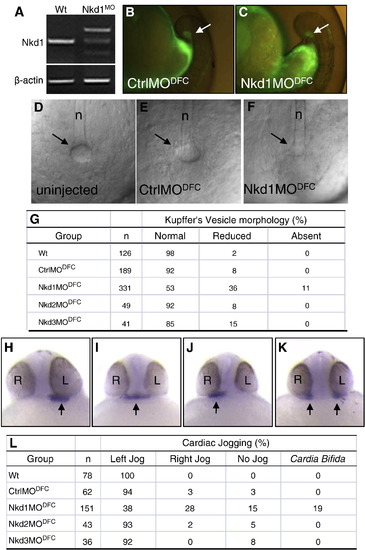
Heart laterality defects after Nkd1 knockdown in DFCs. (A) RT-PCR using cDNA from wt or Nkd1MO-injected embryos: β-actin or Nkd1 were amplified. (B and C) Arrows denote efficient targeting of CtrlMO and Nkd1MO into the DFCs. (D–F) Bright field image of KV in wt (D), CtrlMODFC (E) and Nkd1MODFC(F) embryos, at 10-somite stage. Arrows indicate vesicle location; n = notochord. (G) Summary of KV morphology defects. (H–K) Arrows denote expression of nkx2.5 in the heart, on the left side in wt (H) and on the middle (I), right (J) and bilaterally (K) in Nkd1MODFC embryos. (L) Summary of cardiac jogging defects. EXPRESSION / LABELING:
PHENOTYPE:
|
|
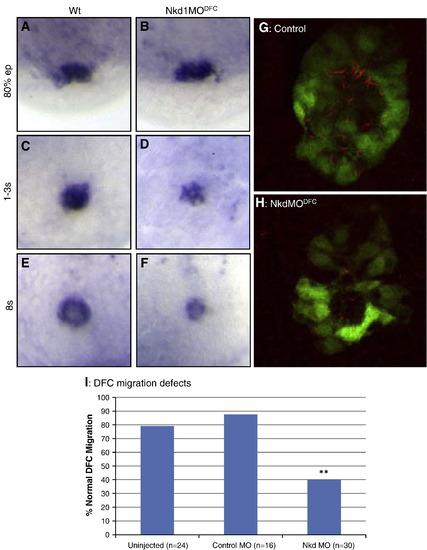
Nkd1 is required for proper DFC migration and KV formation. (A–F) EGFP (Dusp6:d2EGFP) expression in wt (A, C and E) and Nkd1MODFC (B, D and F) embryos; DFCs were assessed at 80% epiboly (A and B), tailbud to 1-somite stage (C and D) and at 8-somite stage (E and F). (G and H) Fluorescence image denoting KV cilia in wt (J) and Nkd1MODFC (K) embryos. Dusp6:d2EGFP (green) and anti-acetylated tubulin (red) indicate cilia in KV cells. (I) Graph of DFC migration defects. N notes the sample size. ** = p < 0.01 compared to uninjected using Fisher′s exact test. EXPRESSION / LABELING:
PHENOTYPE:
|
|
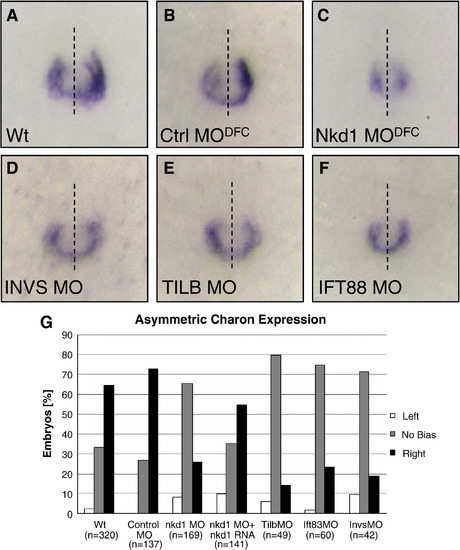
Asymmetric charon expression around the KV is dependent on Nkd1 and cilia function. Expression of charon in wt (A), CtrlMODFC (B), and Nkd1MODFC (C) embryos. Expression of charon in InvsMO (D), SeaMO (E), and Ift88MO (F) injected embryos. Dashed line represents embryonic midline. (G) Graph of percent of embryos displaying left-sided (white bar), no bias (shaded bar) or right-sided biased (black bar) charon expression around KV. (n) notes the sample size for each injection set. ** = p > 0.01 compared to wt using Fisher′s exact test. Nkd1MODFC p-value = 3.07 × 10- 16; Nkd1MODFC+ nkd1 RNA p-value = 0.01, SeaMO p-value = 2.3 × 10- 11; Ift88MO p-value = 3.5 × 10- 9; InvsMO p-value = 2.11 × 10- 8. EXPRESSION / LABELING:
PHENOTYPE:
|
|
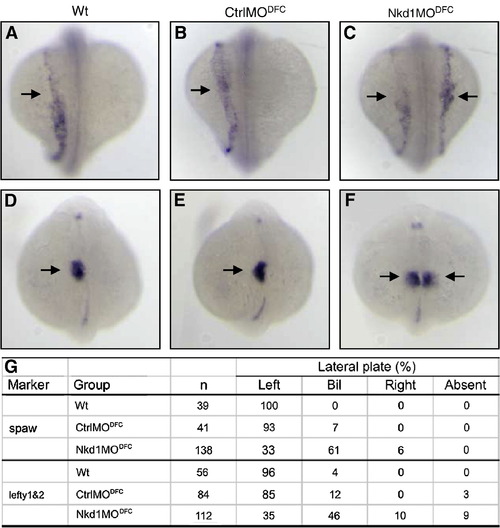
Nkd1 is required for asymmetric expression in the LPM. (A–C) Arrows denote left-sided spaw expression in wt (A), CtrlMODFC (B), and bilateral expression in Nkd1MODFC (C). Arrows denote left-sided lefty1&2 expression in wt (D), CtrlMODFC (E), and bilateral expression in Nkd1MODFC (F). (G) Summary of spaw and lefty1&2 asymmetric gene expression patterns. EXPRESSION / LABELING:
PHENOTYPE:
|
|
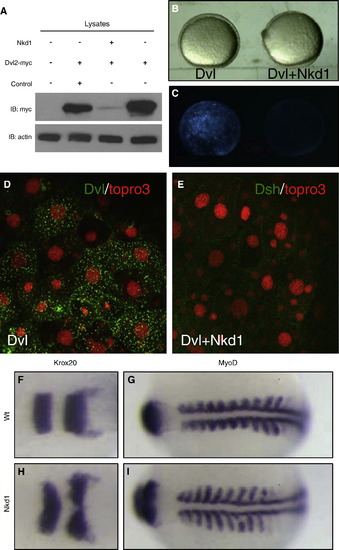
Nkd1 promotes Dvl degradation and impacts CE movements. (A) Westerm blot of zebrafish embryos injected with myc-tagged Dvl2 only, or coinjected with either nkd1 RNA or egfp RNA. The first lane is uninjected embryos, anti-β-actin was used as loading control. Embryos were frozen at 80% epiboly and equal amounts of cell lysates were used for Western blot analysis. (B) Bright field and (C) fluorescence images of embryos at 90% epiboly, injected with Dvl-GFP only (left) or coinjected with nkd1 RNA (right). (D) Dvl-GFP (green) and Topro3 nuclear staining (red) denotes Dvl localization in wt and (E) Dvl-GFP coinjected with nkd1 RNA. (F) krox20 and (G) myoD markers in uninjected and (H) krox20 and (I) myoD markers nkd1 RNA-injected zebrafish embryos. 43% of nkd1 injected embryos show CE defects, n = 143. Dorsal view, anterior to the left. |
|
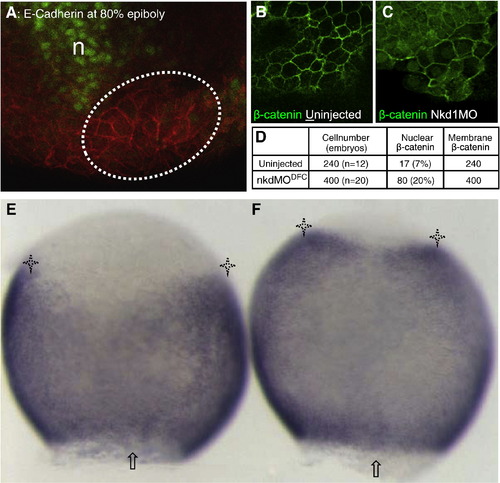
Nkd1 knockdown promotes nuclear β-catenin accumulation in DFCs and activates β-catenin transcriptional targets. Immunohistochemistry on embryos at 80% epiboly. (A) The DFC region (dashed lines) was identified by the position relative to the gsc-GFP positive region (green) and yolk, cell membrane is noted by E-cadherin distribution. (B) In wt DFC, β-catenin is detected mostly in plasma membrane. (C) In Nkd1MODFC embryos, β-catenin is detected at the plasma membrane and in the nuclei of cells in DFC cluster. (D) Summary of β-catenin-positive nuclei present in DFCs of wt and Nkd1MODFC embryos. n = notochord. *P < 6.8 × 10- 6, Fisher′s exact test. WMISH of axin2, dorsal side shown. (E) axin2 expression is excluded from the DFC region (arrow) with distinct lateral domains (asterisks) in ControlMODFC embryos. (F) Expanded axin2 expression is detected in Nkd1MODFC embryos. 91% show expanded domains, n = 33 compared to 7% in control MO, n = 27 (p-value = 2.1 × 10- 11). EXPRESSION / LABELING:
|
|
Nkd1 knockdown promotes β-catenin-dependent transcription. WMISH of TOPdGFP transgenic embryos, dorsal side shown. (A) GFP expression is excluded from the DFC region in ControlMODFC embryos, box notes DFC region magnified in (B). (C) Enhanced GFP expression is detected in Nkd1MODFC embryos, boxed region enlarged in (D). Typical lateral domains of TOPdGFP reporter expression are present in both embryos. 71% of Nkd1MODFC embryos show expanded GFP, n = 28. |
Reprinted from Developmental Biology, 348(1), Schneider, I., Schneider, P.N., Derry, S.W., Lin, S., Barton, L.J., Westfall, T., and Slusarski, D.C., Zebrafish Nkd1 promotes Dvl degradation and is required for left-right patterning, 22-33, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.