- Title
-
WRN loss accelerates abnormal adipocyte metabolism in Werner syndrome
- Authors
- Tian, Y., Lautrup, S., Law, P.W.N., Dinh, N.D., Fang, E.F., Chan, W.Y.
- Source
- Full text @ Cell Biosci.
|
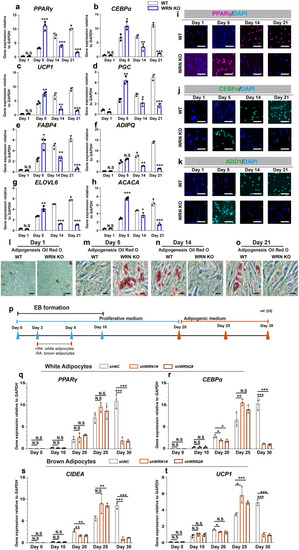
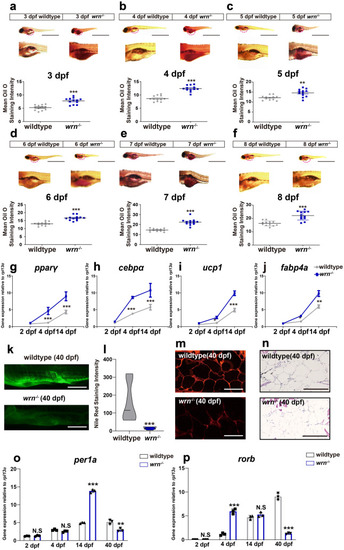
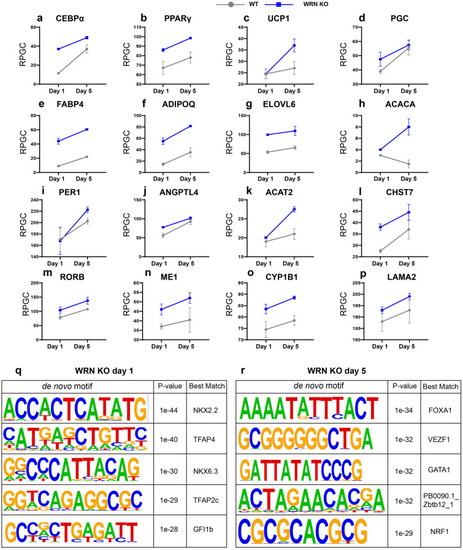
WRN deficiency accelerates adipocyte metabolism. |
|
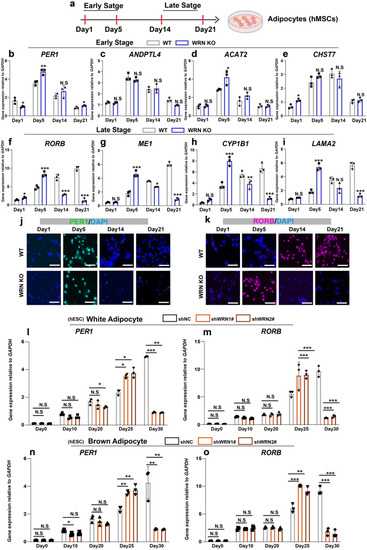
Late-stage adipogenic genes express earlier abnormally. |
|
The EXPRESSION / LABELING:
PHENOTYPE:
|
|
Stage-specific gene regulatory pattern during adipocyte differentiation. |
|
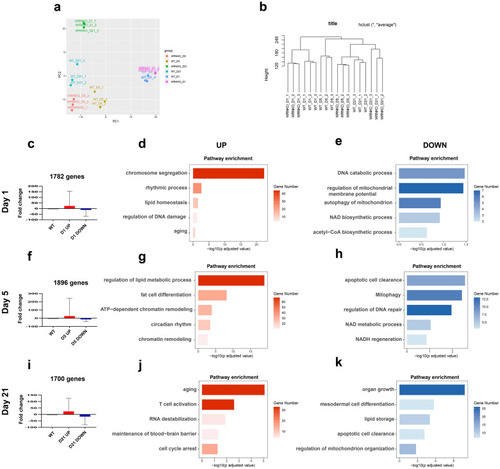
ATAC-seq profiling reflects adipocyte-related chromatin accessibility changes. |
|
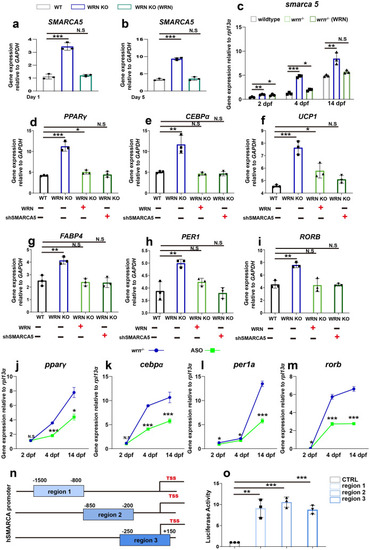
Hyperactive SMARCA5 causes the adipocyte prematurity in WS. EXPRESSION / LABELING:
|
|
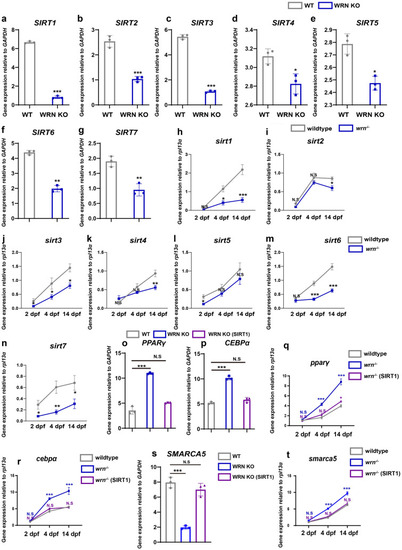
SIRT1 regulates SMARCA5 expression during adipogenesis in hMSCs and zebrafish models. EXPRESSION / LABELING:
PHENOTYPE:
|
|
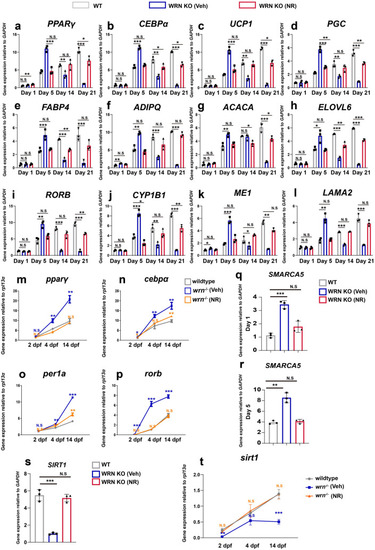
Nicotinamide riboside (NR) normalizes metabolism in WRN KO adipocytes. EXPRESSION / LABELING:
PHENOTYPE:
|