- Title
-
Specific NuRD components are required for fin regeneration in zebrafish
- Authors
- Pfefferli, C., Müller, F., Ja Wi Ska, A., Wicky, C.
- Source
- Full text @ BMC Biol.
|
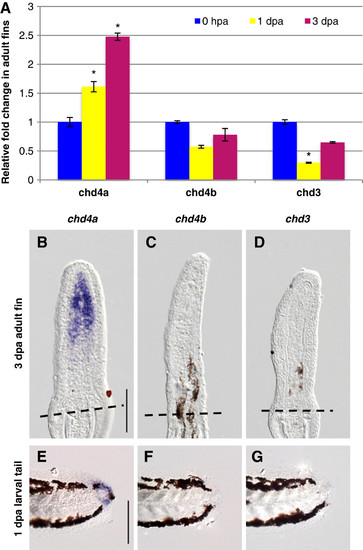
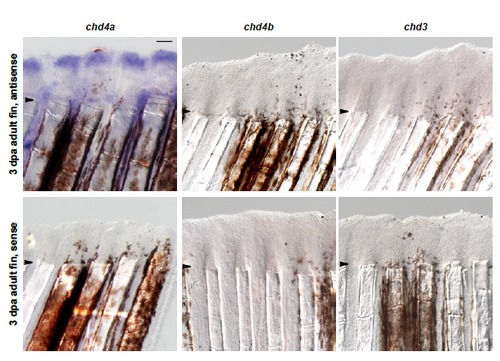
One of the three Mi-2 orthologs, chd4a, is specifically induced in the blastema during fin regeneration. (A) Quantitative real-time-PCR analysis of chd4a, chd4b, and chd3 mRNA in regenerating adult caudal fins at 1 day post-amputation (dpa) and 3 dpa relative to control fins at 0 hours post-amputation (hpa). Error bars represent the SEM. *P < 0.001 (B-D) In situ hybridization with chd4a (B), chd4b (C), and chd3 (D) mRNA antisense probes on cryosections of regenerating adult caudal fins at 3 dpa. Dashed lines indicate the amputation plane. (E-G) Whole-mount in situ hybridization with chd4a (E), chd4b(F), and chd3(G) antisense probes in 1 dpa larval tails. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
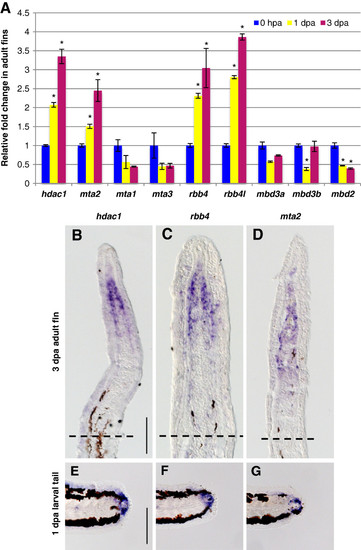
Specific NuRD component orthologs are expressed in the blastema of regenerating fins. (A) Quantitative real-time-PCR analyses of hdac1, mta1, mta2, mta3, rbb4, rbb4l, mbd3a, mbd3b, and mbd2 mRNA in regenerating adult caudal fins at 1 and 3 dpa relative to control fins at 0 hpa. Error bars represent the SEM. *P < 0.001 (B-D) In situ hybridization with hdac1 (B), rbb4(C), and mta2 (D), antisense probes on cryosections of regenerating adult caudal fins at 3 dpa. Dashed lines indicate the amputation plane. (E-G) Whole-mount in situ hybridization with hdac1 (E), rbb4 (F) and mta2 (G) antisense probes in 1 dpa larval tails. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
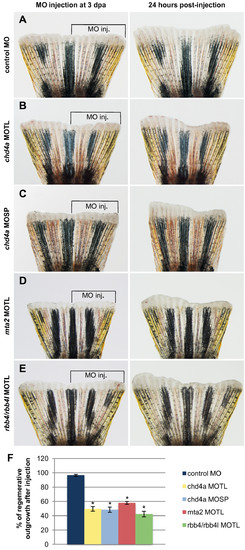
Morpholino-mediated knockdown of chd4a, mta2, and the two RBBP4 orthologs rbb4 and rbb4l results in the reduction in regenerative outgrowth. (A-E) The dorsal half of fins was injected with control (A), chd4a translational blocking (B), chd4a splice blocking (C), mta2 translational blocking (D), and rbb4 + rbb4l translational blocking (E) vivo-morpholinos (MOs). The uninjected ventral half was used as an internal control to assess normal outgrowth. The left panels show fins shortly after injection at 3 dpa and the right panels show the same fins at 24 hours post-injection (hpi). The control MO does not affect regenerative outgrowth compared with the uninjected ventral side of the fins. Injection of the antisense MOs specific for chd4a, mta2, or the two RBBP4 orthologs rbb4 and rbb4l resulted in reduction in regenerative outgrowth compared with the uninjected side. (F) Percentage of regenerative outgrowth after MO injection relative to the uninjected control side of the fins. Error bars represent the SEM. n = 10. *P < 0.01. PHENOTYPE:
|
|
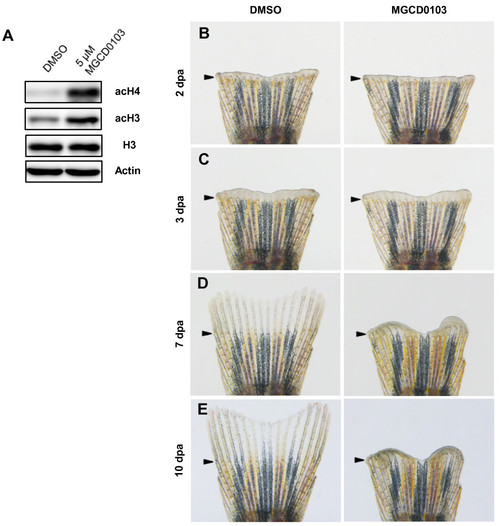
Specific Hdac1 inhibition with MGCD0103 impairs fin regeneration. (A) Western blot analysis for acetylation of histone H3 and histone H4 in 4 dpa fin regenerates treated with DMSO or 5 μM MGCD0103 starting from the time of amputation. The level of acetyl-H3 and acetyl-H4 in MGCD0103-treated fins was increased compared with DMSO-treated fins. β-actin and histone H3 were used as loading controls. (B-E) Whole caudal fins at 2 (B), 3 (C), 7 (D) and 10 dpa (E) treated with DMSO or 5 μM MGCD0103 starting from the time of amputation. MGCD0103 treatment for 10 days impaired regenerative growth without affecting the early stages of regeneration. Arrowheads indicate the amputation plane. PHENOTYPE:
|
|
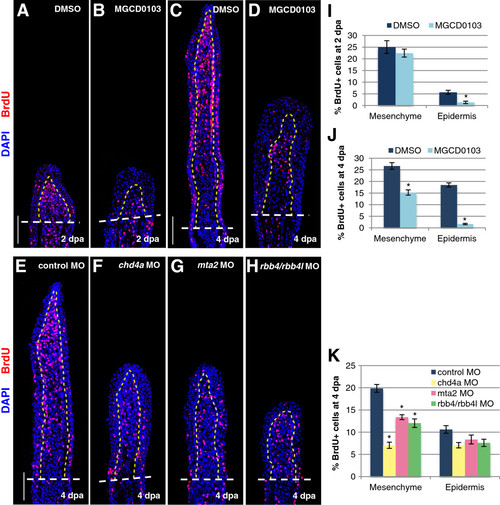
The NuRD components hdac1, chd4a, mta2, and rbb4 are required for blastema cell proliferation during the regenerative outgrowth phase. (A-D) Longitudinal sections of fin regenerates treated with DMSO or MGCD0103 at 2 (A-B) or 4 dpa (C-D) stained with BrdU antibody (red) and DAPI (blue). (E-H) Longitudinal sections of fin regenerates at 4 dpa injected with control (E), chd4a (F), mta2 (G), or rbb4 + rbb4l (H) MOs stained with BrdU antibody (red) and DAPI (blue). Yellow dashed lines indicate the boundary between the blastema and the wound epidermis, and white dashed lines indicate the amputation plane. Scale bars: 100 μm. (I-K) Percentage of BrdU-positive cells relative to the total number of cells (DAPI-labeled) in fin regenerates treated with DMSO or MGCD0103 at 2 dpa (I) or 4 dpa (J) or in fin regenerates injected with control, chd4a, mta2, or rbb4 + rbb4l MOs (K). Error bars represent the SEM. n = 9. *P < 0.01. PHENOTYPE:
|
|
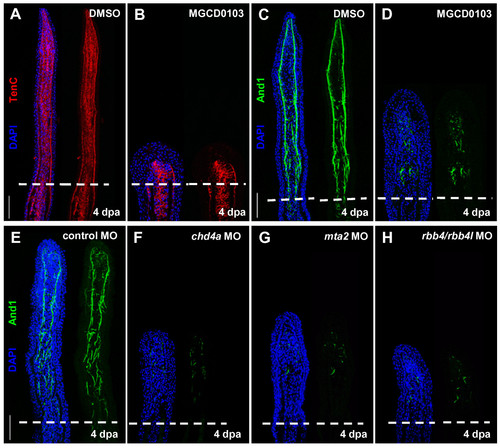
Hdac1 inhibition and morpholino-mediated knockdown of chd4a, mta2, and the two rbb4 orthologs cause abnormal expression of Actinodin 1. (A-B) Longitudinal sections of fin regenerates at 4 dpa treated with DMSO (A) or MGCD0103 (B) and stained with Tenascin C antibody (red) and DAPI (blue). Mesenchymal remodeling was not altered in MGCD0103-treated fins. (C-H) Longitudinal sections of fin regenerates at 4 dpa treated with DMSO (C) or MGCD0103 (D), or injected with control (E), chd4a (F), mta2 (G), or rbb4 + rbb4l (H) MOs stained with Actinodin 1 antibody (green) and DAPI (blue). Depletion of the NuRD components hdac1, chd4a, mta2, or rbb4/rbb4l resulted in a disorganized expression pattern of Actinodin 1. Dashed lines indicate the amputation plane. Scale bars: 100 μm. |
|
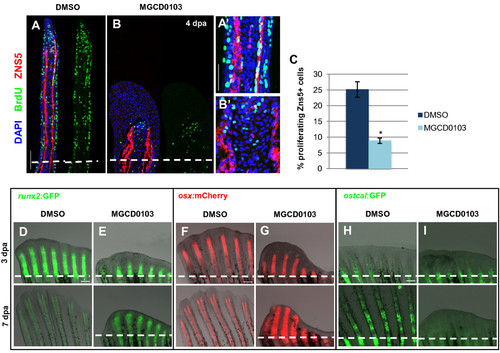
Hdac1 is essential for redifferentiation of osteoblast during regeneration. (A-B) Longitudinal sections of fin regenerates at 4 dpa treated with DMSO (A) or MGCD0103 (B) and triply stained with BrdU (green), ZNS5 antibody (red), and DAPI (blue). Dashed lines indicate the amputation plane. Proximal osteoblast nuclei acquired an elongated shape in control fins (A2), whereas osteoblast nuclei rarely presented an elongated shape in MGCD0103-treated fins (B2). (C) Percentage of ZNS5-positive cells at 4 dpa that were also positive for BrdU relative to the total number of ZNS5-positive cells in fin regenerates treated with DMSO or MGCD0103. Error bars represent the SEM. n = 15. *P < 0.01. (D-I) Caudal fins of runx2:GFP (D-E), osterix:mcherry (F-G) or osteocalcin:GFP (H-I) transgenic fish treated with DMSO or MGCD0103 at 3 and 7 dpa. Constant exposure times were used. Dashed lines indicate the amputation plane. In DMSO-treated fish, the amputation plane is below the photographed part of the fin at 7 dpa. Scale bars: 100 μm (A,D,F,H), 50 μm (A2). |
|
One of the three Mi-2 homologues, chd4a, is induced during regeneration in the adult caudal fin. In situ hybridization with chd4a, chd4b, and chd3 mRNA antisense probes on whole regenerating adult caudal fins at 3 dpa. No signals were detected with the corresponding sense RNA probes. Arrowheads indicate the amputation plane. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
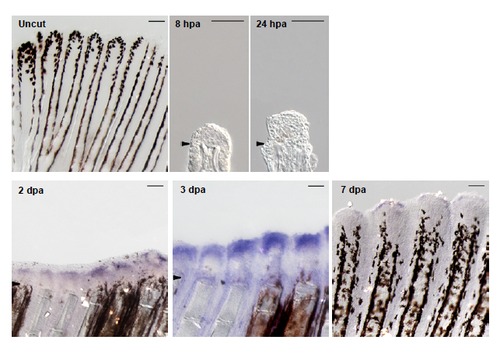
chd4a is expressed during regenerative outgrowth in adult caudal fin. In situ hybridization with chd4a mRNA antisense probe on cryosections at 8 hpa and 24 hpa and on whole fins at 2 dpa, 3 dpa, 7 dpa and in uninjured fin. chd4a expression was first observed at 2 dpa with a robust signal at 3 dpa. Arrowheads indicate the amputation plane. Scale bars: 100 μm. EXPRESSION / LABELING:
|
|
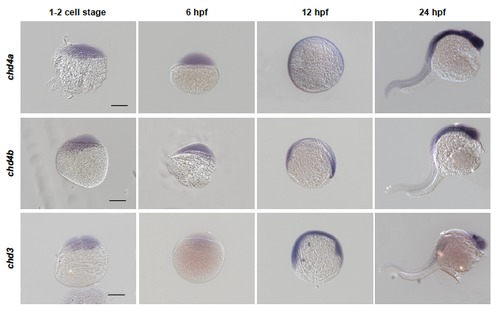
chd4a, chd4b and chd3 are expressed in developing zebrafish embryos. Whole-mount in situ hybridization with chd4a, chd4b and chd3 RNA antisense probes in zebrafish embryos at 1-2 cell stage, 6 hours post-fertilization (hpf), 12 hpf and 24 hpf. The expression pattern of chd4a, chd4b and chd3 in embryos suggest that they might also play a role during embryonic development. Scale bars: 50 μm. |
|
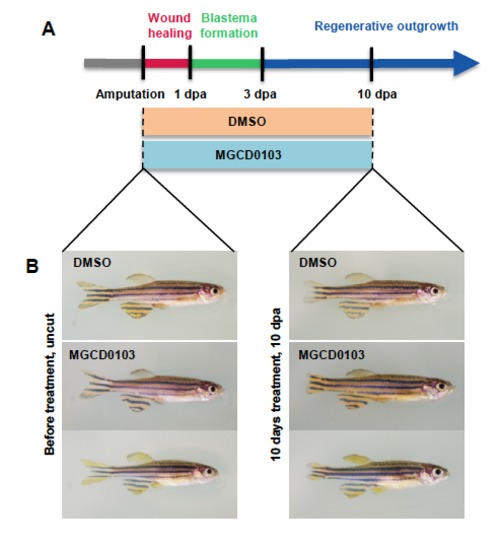
MGCD0103 treatment does not affect the general health of zebrafish. (A) Schematic representation of fin regeneration progression and MGCD0103 treatment. Fish were treated with DMSO or MGCD0103 starting from the time of amputation. (B) Images of fish before and 10 days after DMSO or MGCD0103 treatment. The general health of fish treated with MGCD0103 for 10 days was not obviously altered compared with fish incubated with DMSO-containing water. |
|
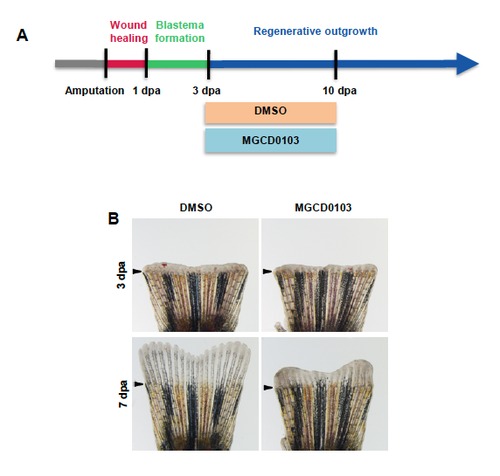
Hdac1 inhibition after blastema formation is sufficient to impair regenerative outgrowth. (A) Schematic representation of fin regeneration progression and MGCD0103 treatment. Fish were treated with DMSO or MGCD0103 starting from 3 dpa. (B) Whole caudal fins at 3 dpa and at 7 dpa treated with DMSO or 5 μM MGCD0103 starting from 3 dpa. Arrowheads indicate the amputation plane. Scale bar: 100 μm. |
|
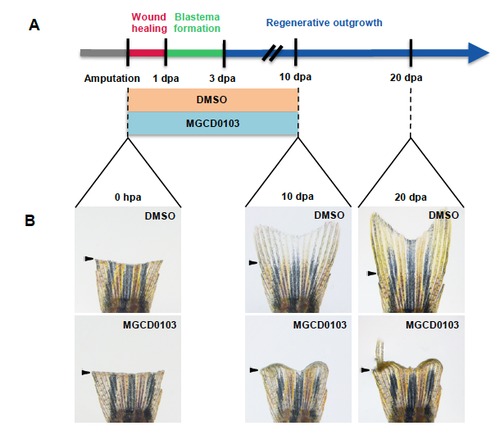
The effects of MGCD0103 treatment are not reversible. (A) Schematic representation of fin regeneration progression and MGCD0103 treatment. Fish were treated with DMSO or MGCD0103 for 10 days starting from the time of amputation and then fish were transferred into normal fish water for 10 additional days. (B) Corresponding images of whole caudal fins just after amputation (0 hpa), at 10 dpa or at 20 dpa. Effects of MGCD0103 treatment were not reversible. In some cases, a few rays were able to resume the regeneration process. Arrowheads indicate the amputation plane. |
|
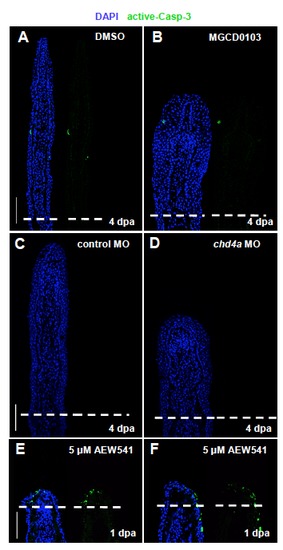
Hdac1 inhibition and chd4a knockdown do not result in the activation of the apoptosis marker caspase-3. (A-F) Longitudinal sections of fin regenerates stained with active caspase-3 antibody (green) and with DAPI (blue). Only a few cells positive for active caspase-3 are present in control fins (A,C). Fins treated with MGCD0103 (B) or injected with chd4a MO (D) do not display any increase of apoptosis at 4 dpa. (E-F) As a positive control, the activation of caspase-3 was assessed in fin regenerates at 1 dpa treated with the chemical inhibitor of Igf1 receptor NVP_AEW541. Inhibition of IGF signaling blocks fin regeneration and results in the increase of cell apoptosis in the wound epidermis (Chablais et al., 2010). Dashed lines indicate the amputation plane. Scale bars: 100 μm. PHENOTYPE:
|
|
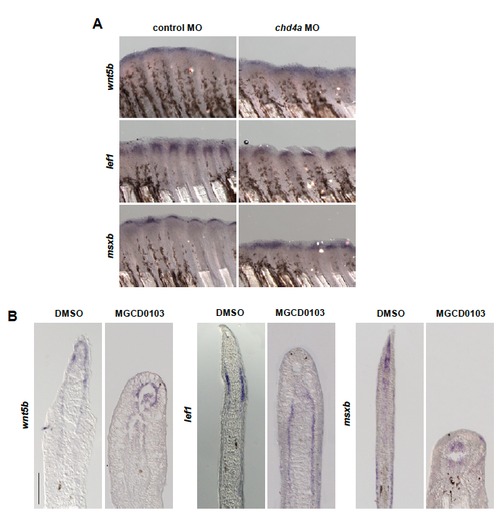
wnt5b, lef1 and msxb are expressed in chd4a MO-injected and MGCD0103-treated fins. (A-B) In situ hybridization with wnt5b, lef1 and msxb mRNA antisense probes in chd4a MO-injected (A) and in MGCD0103-treated (B) fin regenerates at 4 dpa. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
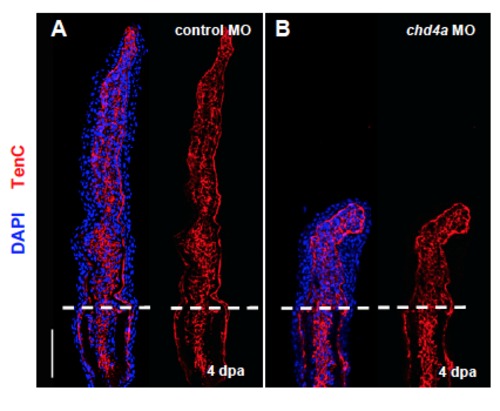
chd4a knockdown does not affect Tenascin C expression. (A-B) Longitudinal sections of fin regenerates at 4 dpa injected with control (A) or chd4a (B) MOs stained with Tenascin C antibody (red) and with DAPI (blue). Mesenchymal remodeling was not altered in chd4a MO-injected fins. Dashed lines indicate the amputation plane. Scale bars: 100 μm. EXPRESSION / LABELING:
|
|
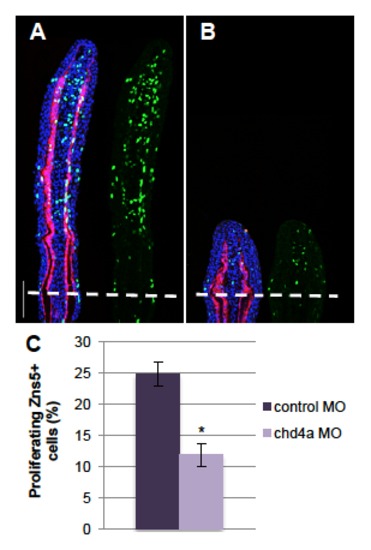
chd4a knockdown reduces osteoblast proliferation. (A-B) Longitudinal sections of fin regenerates at 4 dpa injected with control (A) or chd4a (B) MOs triply stained with BrdU (green), ZNS5 antibody (red) and DAPI (blue). Dashed lines indicate the amputation plane. Scale bar: 100 μm. (C) Percentage of ZNS5-positive cells that were also positive for BrdU relative to the total number of ZNS5-positive cells in fin regenerates at 4 dpa injected with with control or chd4a MOs. Error bars represent the s.e.m. n = 17. * P<0.01. PHENOTYPE:
|
|
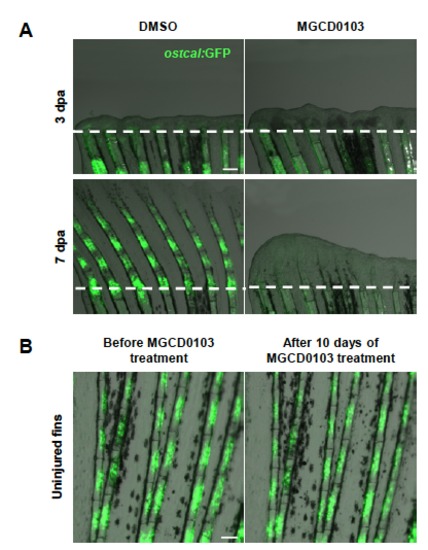
Hdac1 inhibition after blastema formation is sufficient to block reactivation of osteocalcin expression. (A) Caudal fins of osteocalcin:GFP transgenic fish at 3 dpa and 7 dpa treated with DMSO or MGCD0103 starting at 3 dpa. osteocalcin:GFP expression was not reactivated in fin regenerates treated with MGCD0103 after blastema has been formed. (B) Uninjured caudal fins of osteocalcin:GFP transgenic fish before and 10 days after MGCD0103 treatment. MGCD0103 treatment does not alter osteocalcin:GFP expression in uninjured fins. Constant exposure times were used. Scale bars: 100 μm. |