- Title
-
Zebrafish tsc1 and cxcl12a increase susceptibility to mycobacterial infection
- Authors
- Wright, K., Han, D.J., Song, R., de Silva, K., Plain, K.M., Purdie, A.C., Shepherd, A., Chin, M., Hortle, E., Wong, J.J., Britton, W.J., Oehlers, S.H.
- Source
- Full text @ Life Sci Alliance
|
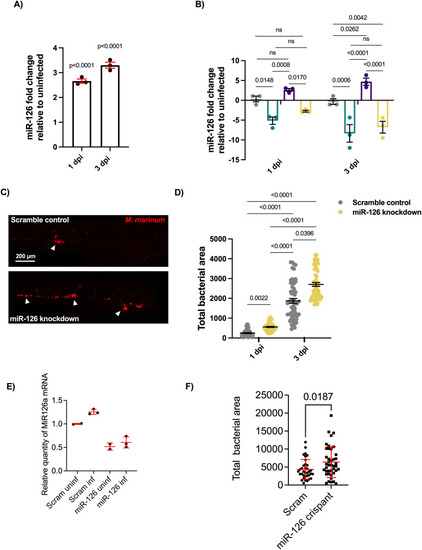
Infection-induced miR-126 expression alters bacterial burden. |
|
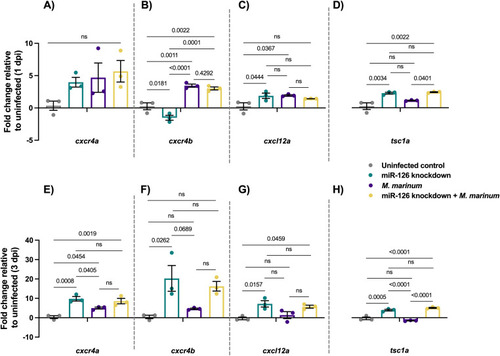
Expression of potential miR-126 mRNA targets is conserved in zebrafish |
|
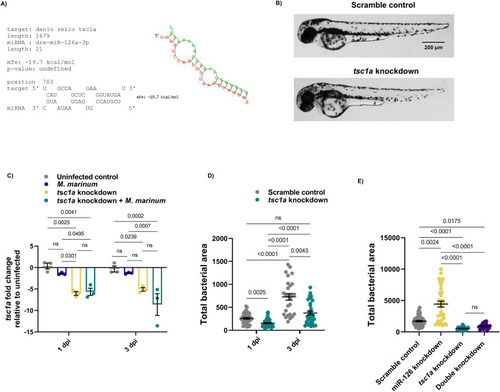
miR-126 potentially targets |
|
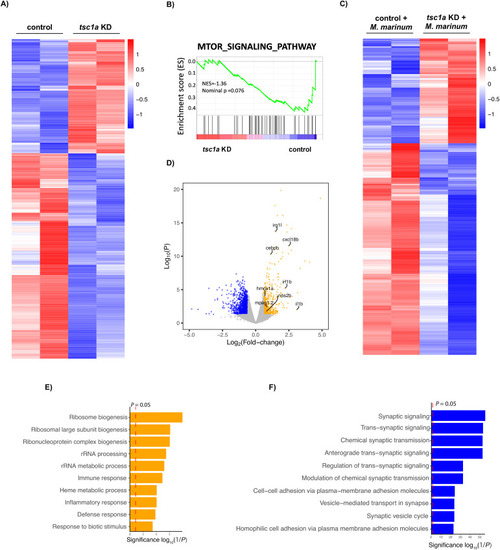
Modulation of Tsc1 alters key host defence and inflammatory pathways. |
|
miR-126 acts on |
|
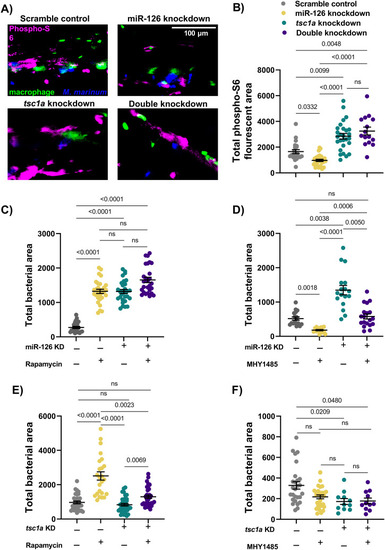
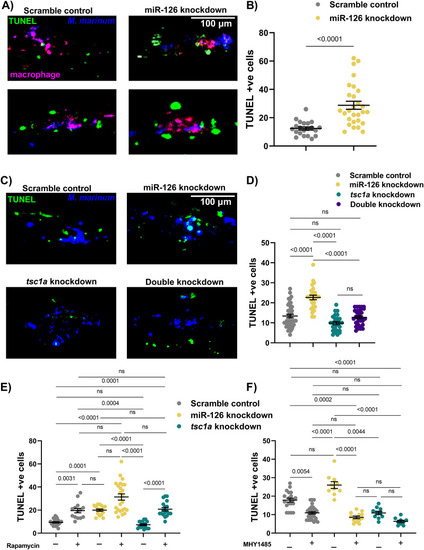
Decreased mTOR signalling alters cell death dynamics in mycobacterial infection. |
|
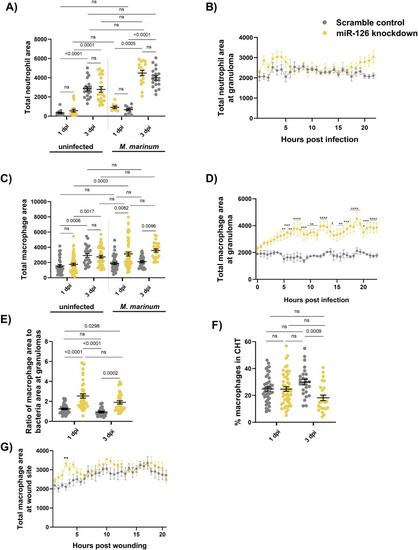
Mycobacterial infection-induced miR-126 expression alters the host macrophage response. |
|
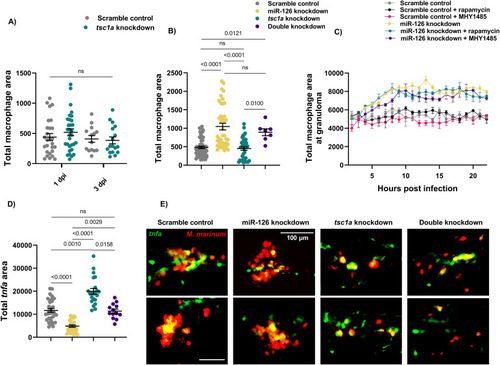
miR-126–dependent macrophage responses to infection are not controlled by the Tsc1a/mTOR signalling axis. |
|
Mycobacterial infection-induced miR-126 expression increases proinflammatory bactericidal macrophage recruitment. |
|
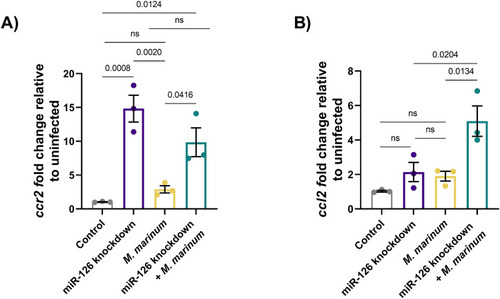
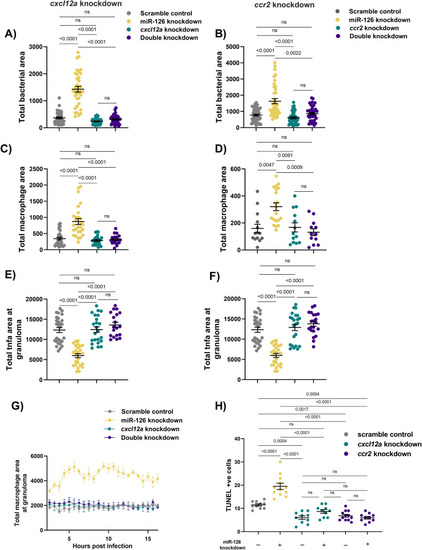
Infection-induced miR-126 regulates Cxcl12/Ccl2/Ccr2 signalling to restrict macrophage recruitment to sites of infection. |
|
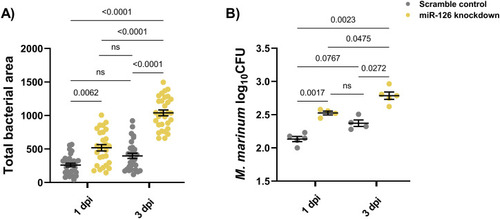
Comparison of infectious burden measurements in scramble control and miR-126 knockdown embryos at 1 and 3 dpi. |
|
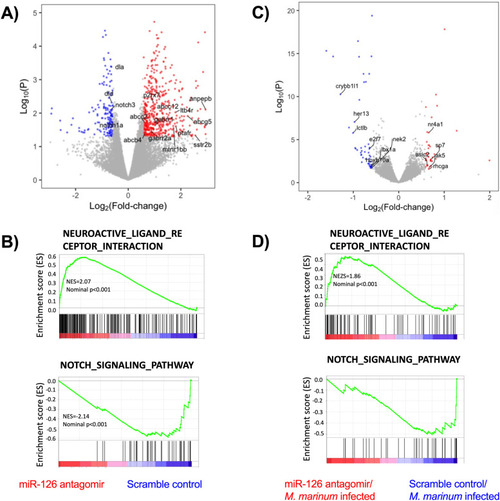
Modulation of miR-126 alters neural and notch signaling pathways. |
|
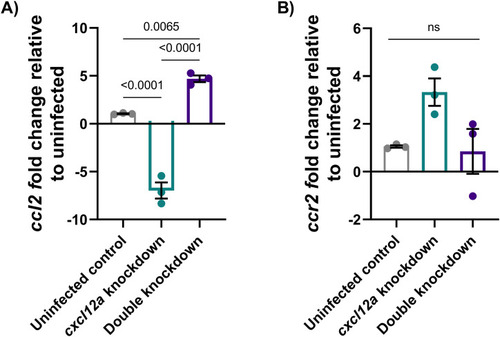
ccl2 but not ccr2 expression is dependent on cxcl12a. |