- Title
-
Hand2 Regulates Extracellular Matrix Remodeling Essential for Gut-Looping Morphogenesis in Zebrafish
- Authors
- Yin, C., Kikuchi, K., Hochgreb, T., Poss, K.D., and Stainier, D.Y.
- Source
- Full text @ Dev. Cell
|
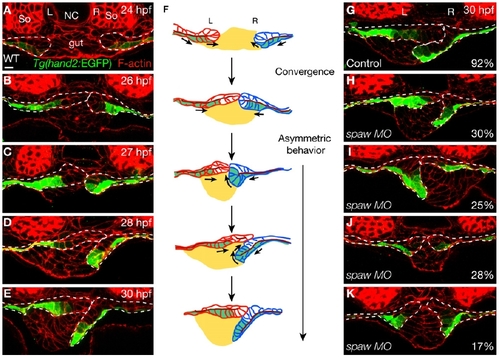
Examination of Tg(hand2:EGFP) Embryos Reveals Novel Cell Rearrangements during LPM Migration (A–E) Time course analysis of Tg(hand2:EGFP) expression in the LPM during gut looping. Ten Tg(hand2:EGFP) embryos were fixed every hour between 24 and 30 hpf, and stained for GFP (green) and phalloidin (red). (F) Diagram of LPM migration. The gut is in yellow, the left LPM is in red, the right LPM is in blue, and the Tg(hand2:EGFP)-expressing cells are in green. The dark red line represents the apical side of the LPM epithelium. Arrows indicate the direction of migration of the Tg(hand2:EGFP)-expressing cells. (G–K) Perturbing spaw function randomized the laterality of the rearrangements of the Tg(hand2:EGFP)-expressing cells. Compared to uninjected controls (G; n = 12), of 53 embryos injected with 10 ng of spaw MO, 30% showed the wild-type pattern (H); 25% showed the reverse pattern (I); 28% had both the left and right LPM migrate on top of the gut while the Tg(hand2:EGFP)-expressing cells on both sides remained ventral (J); and 17% had both the left and right LPM migrate ventrally and the Tg(hand2:EGFP)-expressing cells on both sides “rolled” dorsally (K). All images are transverse sections, dorsal to the top. Dashed lines outline the LPM. L, left; NC, notochord; R, right; So, somite. The scale bar represents 10 μm. |
|
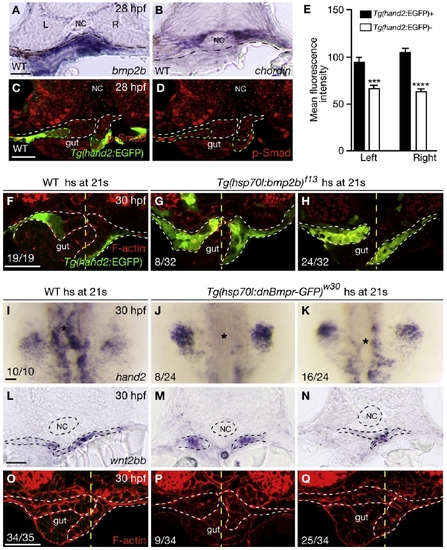
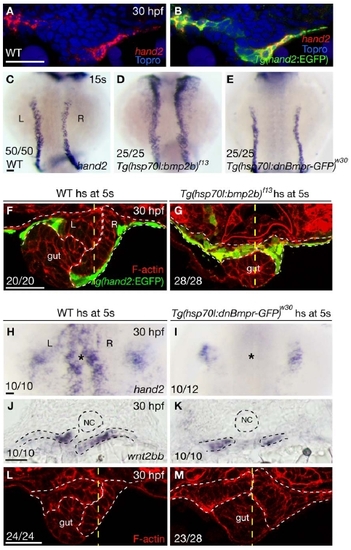
Bmp Signaling Regulates hand2 Expression in the LPM (A and B) Within the gut-looping region of wild-type embryos, bmp2b (A) is expressed in the LPM, whereas chordin (B) is expressed in the ventral portion of the somites. Dashed lines mark the somite/LPM boundary. (C) Wild-type embryo stained for GFP (green) and phosphorylated Smad1/5/8 (red). (D) The same embryo as shown in (C) but with red channel only. Dashed lines outline the LPM. (E) Fluorescence intensity of the phosphorylated Smad staining in the LPM cells (mean ± SEM). Forty Tg(hand2:EGFP)-expressing cells and 40 Tg(hand2:EGFP)-nonexpressing cells from seven wild-type embryos were analyzed. Asterisks indicate statistical significance: ***p < 0.001; ****p < 0.0001. (F–H) Wild-type and Tg(hsp70l:bmp2b)f13 embryos were heat shocked at the 21-somite stage, and stained for GFP (green) and phalloidin (red) at 30 hpf. White dashed lines outline the LPM. Yellow dashed lines mark the midline. Tg(hand2:EGFP) was expressed mostly in the ventral half of the LPM in wild-types (F), but was expressed throughout the LPM in the gut-looping region in Tg(hsp70l:bmp2b)f13 embryos (G and H). Eight of 32 Tg(hsp70l:bmp2b)f13 embryos examined exhibited no gut looping (G); the remaining 24 exhibited leftward gut looping (H). (I–K) Compared to wild-type control (I), hand2 expression in the LPM (asterisks) was decreased (K) or absent (J) in Tg(hsp70l:dnBmpr-GFP)w30 embryos after heat shock. (L–N) Expression of wnt2bb in the LPM was present in wild-type and Tg(hsp70l:dnBmpr-GFP)w30 embryos after heat shock. Dashed lines outline the notochord and LPM. (O–Q) Phalloidin staining of wild-type control (O) and Tg(hsp70l:dnBmpr-GFP)w30 (P and Q) embryos. When heat shock was applied at the 21-somite stage, 9 of 34 embryos exhibited no gut looping (P); the remaining 25 exhibited leftward gut looping (Q). White dashed lines outline the LPM. Yellow dashed lines mark the midline. All images, except (I)–(K), are transverse sections, dorsal to the top. (I)–(K) are dorsal views, anterior to the top. L, left; NC, notochord; R, right. The scale bars represent 40 μm. See also Figure S1. |
|
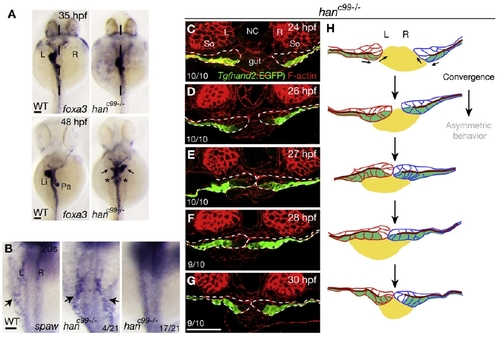
Hand2 Is Required for Gut Looping and LPM Migration (A) foxa3 expression in wild-type and hanc99 mutants. Upper panel: the gut has looped to the left by 35 hpf in wild-types (22/22), but stays in the midline in hanc99 mutants (17/18). Dashed lines mark the midline. Lower panel: in wild-type by 48 hpf, the liver and pancreas are positioned on the left and right side of the gut, respectively (35/35). In hanc99 mutants, the liver (arrows) and/or pancreas (asterisks) is duplicated (12/12). (B) At the 20-somite stage (19 hpf), spaw is expressed in the left LPM (arrows) in wild-types (70/70), but is absent from the LPM in most hanc99 mutants, or is bilateral in a few hanc99 mutants. (C–G) Time course analysis shows that the Tg(hand2:EGFP)-expressing cells in hanc99 mutants fail to undergo asymmetric rearrangements. Ten hanc99 mutants were analyzed at each time point. Dashed lines outline the LPM. (H) Diagram of LPM migration in hanc99 mutants. The gut is in yellow, the left LPM is in red, the right LPM is in blue, and the Tg(hand2:EGFP)-expressing cells are in green. The dark red line represents the apical side of the LPM epithelium. Arrows indicate the direction of migration of the Tg(hand2:EGFP)-expressing cells. (A and B) Dorsal views, anterior to the top. (C–H) Transverse sections, dorsal to the top. L, left; Li, liver; NC, notochord; Pa, pancreas; R, right; So, somite. The scale bars represent 50 μm. See also Figure S2. EXPRESSION / LABELING:
|
|
hanc99 Mutants Exhibit Prolonged Deposition of Laminin at the LPM/Gut Boundary (A–F) Time course analysis of Tg(hand2:EGFP) expression and laminin deposition during LPM migration. Ten wild-type (A–C) and eight hanc99 mutants (D–F) were fixed at the stages indicated and stained for laminin (red) and GFP (green). Dashed lines outline the notochord. (A–C) In wild-type, laminin becomes diminished along the migratory path of Tg(hand2:EGFP)-expressing cells (arrows). (D–F) In hanc99 mutants, laminin deposition persists along the entire LPM/gut boundary (arrows). (G) The LPM in lamβ1s804 heterozygotes undergoes asymmetric migration (28/30). (H–J) Out of 29 lamβ1s804 mutants examined, 6 showed incomplete asymmetric LPM migration (H), 18 had both the left and right LPM remain on top of the gut (I), and 5 had the LPM located lateral to the gut (J). Arrows point to the aberrant intrusion of LPM cells into the gut. Dashed lines outline the LPM. (K) Percentages (mean ± SEM) of hanc99 mutants (n = 21) from hanc99+/- incrosses and hanc99 single mutants (n = 56) from hanc99+/-;lamβ1s804+/- incrosses showing gut-looping phenotypes according to foxa3 expression. Three independent crosses were examined. All images are transverse sections, dorsal to the top. L, left; NC, notochord; R, right. The scale bars represent 40 μm. See also Figure S3. |
|
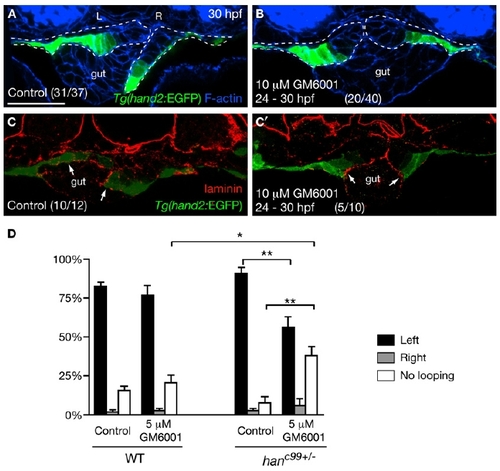
Asymmetric LPM Migration Requires the Diminishment of Laminin by MMPs (A and B) MMP inhibition blocked the asymmetric LPM migration. Tg(hand2:EGFP) embryos were treated with DMSO (A) or 10 μM GM6001 (B) from 24 hpf, and stained for GFP (green) and phalloidin (blue) at 30 hpf. Dashed lines outline the LPM. (C and C′) Laminin (red) deposition was diminished near the Tg(hand2:EGFP)-expressing cells in the DMSO-treated controls (C, arrows), but persisted along the entire LPM/gut boundary in GM6001-treated embryos (C′, arrows). (D) Proportions (mean ± SEM) of wild-type and hanc99+/- embryos showing gut-looping phenotypes according to foxa3 expression. Three independent experiments were conducted. Two hundred wild-type and 200 genotyped hanc99+/- embryos treated with DMSO or 5 μM GM6001 were examined. Asterisks indicate statistical significance: *p < 0.05; **p < 0.01. (A–C′) Transverse sections, dorsal to the top. L, left; R, right. The scale bar represents 40 μm. See also Figure S2. |
|
Loss of Hand2 Function Reduces MMP Activity along the LPM/Gut Boundary (A) Diagram showing the microinjection of the DQ-collagen IV substrate into the embryo. Lateral view, anterior to the left. The blue dot marks the position where gut looping occurs. (B) Tg(hand2:EGFP) embryos were injected with DQ-collagen IV at 26 hpf, fixed at 27 hpf, and stained for GFP (blue) and phalloidin (red). The fluorescein signal caused by cleavage of the DQ-collagen substrate is shown in green. (C) In embryos preinjected with GM6001, almost no fluorescein signal could be detected. (D and E) Wild-type (D) and hanc99-/- (E) embryos were coinjected with DQ-collagen and Alex549-conjugated collagen at 27 hpf and fixed at 28 hpf. The spots where the fluorescence intensity was measured are marked by pink and orange bars. (F) MMP activity represented as normalized fluorescence intensity (mean ± SEM). Six wild-type and six hanc99 mutants were examined at each time point. Asterisks indicate statistical significance: *p < 0.05. (B–E) Transverse sections, dorsal to the top. L, left; NC, notochord; NT, neural tube; R, right; So, somite. The scale bars represent 40 μm. |
|
Expression of mmp Genes and Their Inhibitors Is Altered in hand2 Mutants (A) Comparison of mmp and timp expression in wild-type and hans40-/- samples by quantitative real-time PCR (two independent experiments, three replicates of wild-type and hans40-/- samples each time). The results are represented as relative expression levels that are normalized to β-actin (mean ± SEM). (B) Proportion (mean ± SEM) of wild-type embryos (n = 29), wild-type embryos injected with 0.5 ng of mmp14a MO (n = 44), hanc99 heterozygotes (n = 55), and hanc99 heterozygotes injected with 0.5 ng of mmp14a MO (n = 70) showing gut-looping phenotypes according to foxa3 expression. Three independent experiments were conducted. (A and B) Asterisks indicate statistical significance: *p < 0.05; **p < 0.01. (C) Diagram of Hand2 regulating asymmetric LPM migration during zebrafish gut-looping morphogenesis. See also Figure S4. |
|
Bmp signaling regulates hand2 expression in the lateral plate mesoderm (LPM) |
|
The LPM in the gut-looping region of hanc99 mutants does not exhibit impaired epithelial polarity or altered Fibronectin deposition |
|
Expression levels of laminin and integrin genes are not dramatically altered in hans40 mutants compared to wild-type. |
|
Expression of mmp14a, timp2a, and timp2b in wild-type and hanc99-/- embryos at 28 hpf For each gene, 10 wild-type and 10 hanc99-/- embryos were examined. (A-F) are dorsal views, anterior to the top. Squares mark the gut-looping region. (A′-F′) are transverse sections, dorsal to the top. Dashed lines outline the notochord and the LPM. L, left; NC, notochord; R, right. Scale bars, 40 μm. |

Unillustrated author statements |
Reprinted from Developmental Cell, 18(6), Yin, C., Kikuchi, K., Hochgreb, T., Poss, K.D., and Stainier, D.Y., Hand2 Regulates Extracellular Matrix Remodeling Essential for Gut-Looping Morphogenesis in Zebrafish, 973-984, Copyright (2010) with permission from Elsevier. Full text @ Dev. Cell