- Title
-
Naringin ameliorates liver fibrosis in zebrafish by modulating IDO1-mediated lipid metabolism and inflammatory infiltration
- Authors
- Qin, M.C., Li, J.J., Zheng, Y.T., Li, Y.J., Zhang, Y.X., Ou, R.X., He, W.Y., Zhao, J.M., Liu, S.T., Liu, M.H., Lin, H.Y., Gao, L.
- Source
- Full text @ Food Funct
|
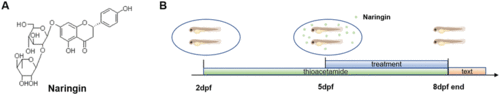
Chemical structure of naringin and the schematic diagram of experimental protocols. (A) Chemical structure of naringin. (B) Schematic diagram of the experimental protocols in zebrafish |
|
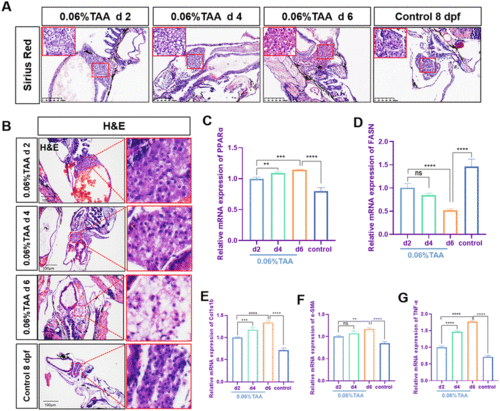
Establishment of TAA-induced liver fibrosis model in zebrafish. (A) The Sirius red staining of zebrafish. Figures are magnified at ×100 (n = 8). (B) H&E staining of zebrafish. Figures are magnified at ×100 (n = 8). (C–G) The qPCR analysis of PPARα, FASN, Col1α1b, α-SMA, and TNF-α mRNA expression in zebrafish larvae (n = 3 or 4). The control group consisted of untreated zebrafish at 8 dpf. The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
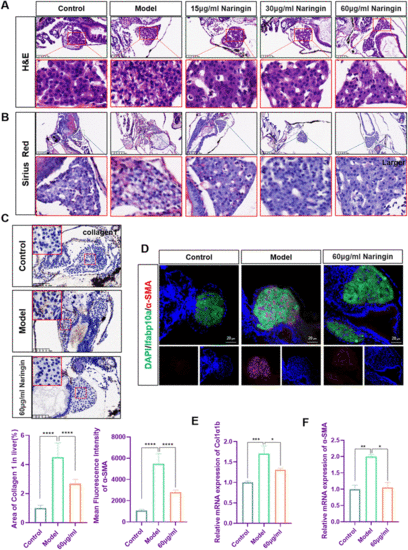
Naringin attenuated TAA-induced liver fibrosis in zebrafish. (A) H&E staining of zebrafish larvae. Figures are magnified as ×200 (n = 8). (B) Sirius red staining of zebrafish larvae. Figures are magnified at ×100 (n = 8). (C) Immunohistochemical staining of collagen1 in paraffin sections of zebrafish larvae. Figures are magnified at ×200 (n = 8). (D) Frozen liver sections of zebrafish larvae with liver-specific eGFP expression were immunofluorescently stained with α-SMA (n = 7). (E) The qPCR analysis of Col1a1b mRNA expression in zebrafish (n = 3). (F) The qPCR analysis of α-SMA mRNA expression in zebrafish (n = 3). The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
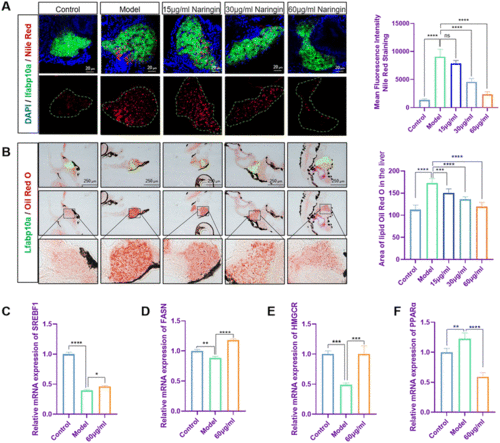
Naringin alleviated lipid metabolism disorders in TAA-induced zebrafish liver fibrosis. (A) Nile red staining of frozen sections of zebrafish larvae (n = 7). (B) Oil red O staining of zebrafish larvae. Figures are magnified at ×100 (n = 7). (C–F) The qPCR analysis of SREBF1, FASN, HMGCR and PPARα mRNA expression in zebrafish larvae (n = 3). The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
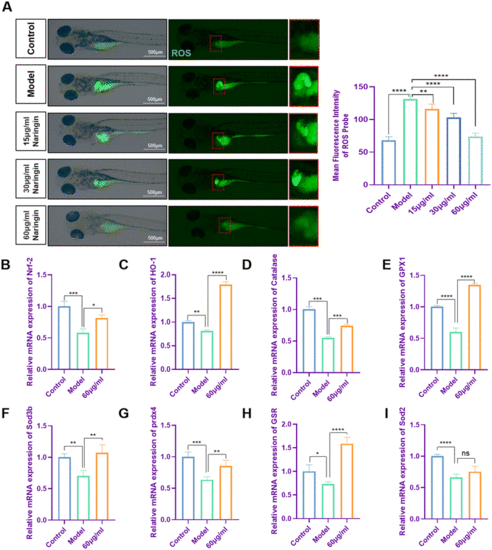
Naringin decreased oxidative stress in zebrafish liver fibrosis. (A) Fluorescence micrographs of DCFH-DA shown in green. Figures are magnified at 32× (n = 8). (B–I) The qPCR analysis of Nrf-2, HO-1, GPX1, catalase, Sod3b, Prdx4, GSR, and Sod2 mRNA expression in zebrafish larvae (n = 3 or 4). The control group consisted of untreated zebrafish at 8 dpf. The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denoted no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
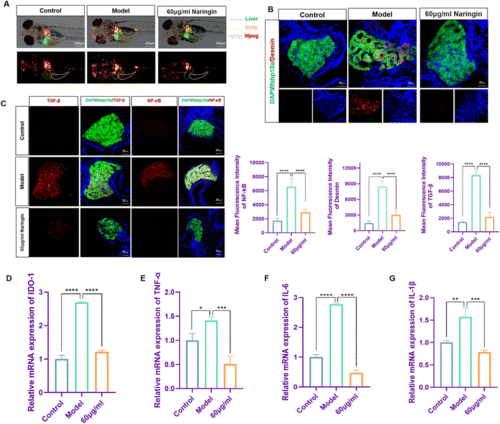
Naringin reduced activation of inflammation and HSCs in TAA-induced liver fibrosis in zebrafish. (A) Naringin reduced the infiltration of macrophages in the alimentary canal of zebrafish during TAA exposure. Figures are magnified at 40× (n = 8). (B) Frozen liver sections of zebrafish larvae with liver-specific eGFP expression were immunofluorescently stained with desmin (n = 7). (C) Frozen liver sections of zebrafish larvae with liver-specific eGFP expression were immunofluorescently stained with TGF-β and NF-κB (n = 7). (D–G) The qPCR analysis of IDO1, TNF-α, IL-6, and IL-1β mRNA expression in zebrafish larvae (n = 3). The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
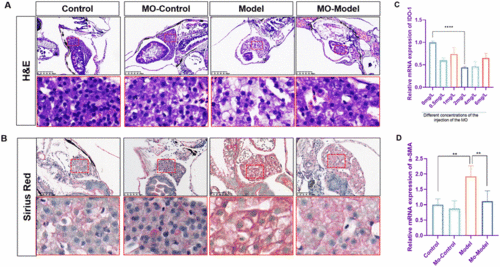
Knock-down of IDO1 reversed the fibrosis degree in zebrafish. (A) H&E staining of zebrafish larvae. Figures are magnified at ×200 (n = 8). (B) Sirius red staining of zebrafish larvae. Figures are magnified at ×200 (n = 8) (C and D) The qPCR analysis of IDO1 and α-SMA mRNA expression in zebrafish larvae (n = 3). The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
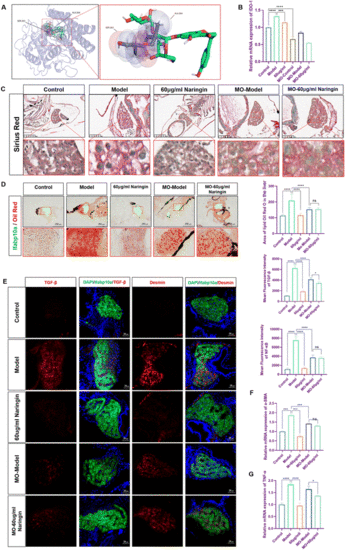
Knock-down of IDO1 abolished naringin-mediated suppression of fibrosis in zebrafish. (A) Molecular docking of Naringin and IDO1. (B) The qPCR analysis of IDO1 mRNA expression in zebrafish larvae (n = 3). (C) Sirius red staining of zebrafish larvae. Figures are magnified at ×200 (n = 8). (D) Oil red O staining of zebrafish larvae. Figures are magnified at ×100 (n = 8). (E) Frozen liver sections of zebrafish larvae with liver-specific eGFP expression were immunofluorescently stained with TGF-β and Desmin (n = 7). (F and G) The qPCR analysis of α-SMA and TNF-α mRNA expression in zebrafish larvae (n = 3). The mRNA expression was normalized to β-actin mRNA expression and presented as a fold change compared with the control group. ns denotes no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. |
|
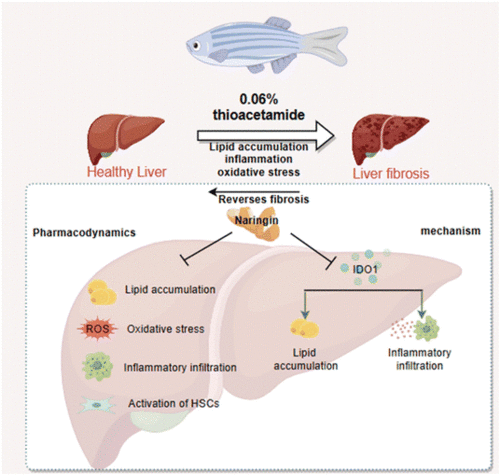
Diagram of the protective mechanism of naringin on LF. |