- Title
-
Stochastic gene expression and environmental stressors trigger variable somite segmentation phenotypes
- Authors
- Keseroglu, K., Zinani, O.Q.H., Keskin, S., Seawall, H., Alpay, E.E., Özbudak, E.M.
- Source
- Full text @ Nat. Commun.
|
Segmentation defects in mutants are stochastic. |
|
Gene expression in mutants are stochastic. |
|
An amplitude threshold of oscillations is required for successful segmentation in a genetic background displaying variable expressivity. |
|
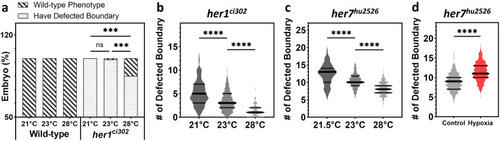
Environmental factors affect the strength of segmentation phenotypes. |
|
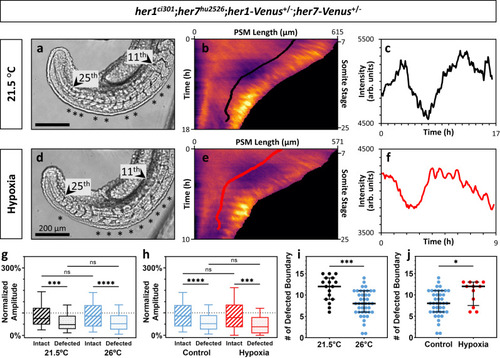
Segmentation clock amplitudes correlate with segmentation phenotypes affected by environmental factors. |
|
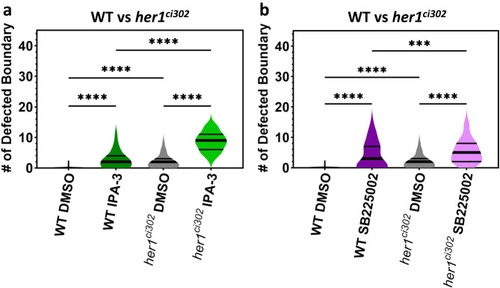
Embryotoxic chemicals worsen segmentation defects. |