- Title
-
Pnrc2 regulates 3'UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation
- Authors
- Gallagher, T.L., Tietz, K.T., Morrow, Z.T., McCammon, J.M., Goldrich, M.L., Derr, N.L., Amacher, S.L.
- Source
- Full text @ Dev. Biol.
|
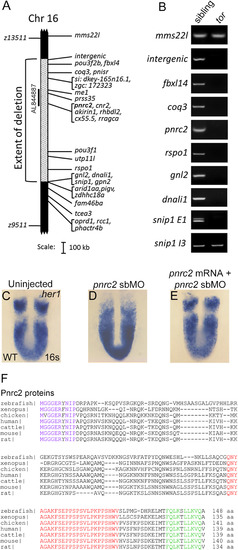
The tortugab644 allele is a 1.46 Mb deficiency that includes the pnrc2 gene. Haploid-based mapping revealed that the tortuga lesion lies 0.30 cM to the left (4/1434 recombinants) and 0.37 cM to the right (4/1083 recombinants) of the SSLP markers z13511 and z9511, respectively (A). The extent of the deletion was refined by PCR-based screening of regions within and around the deletion interval from pooled genomic DNA samples of 10 wild-type (WT) and 10 tortuga mutant embryos (B); regions that fail to amplify in mutants are deleted in the torb644 allele (A, B). Among BACs spanning the deletion interval, only BAC AL844887 restores her1 expression when injected into tor mutants (A; Fig S1A-D). In wild-type embryos, her1 is expressed in a striped pattern (n=54/54) (C). In contrast, embryos injected with 6 ng of pnrc2 splice-blocking morpholino (sbMO) have a tortuga-like her1 expression defect (n=37/39) (D). When pnrc2 mRNA is co-injected with 4 ng pnrc2 sbMO, her1 expression is partially restored in a dose-dependent manner (n=3/19 WT her1 expression, 150 pg pnrc2 mRNA; n=7/17 WT her1 expression, 600 pg pnrc2 mRNA) (E; Table 1). Alignment of vertebrate Pnrc2 amino acid sequences reveals a conserved 10 amino acid N-terminus (purple) and conserved C-terminal SRC-Homology 3 (SH3) (red) and Nuclear Receptor (NR box) domains (green) (F). Clustal alignments were performed in consultation with published alignments for vertebrate Pnrc2 (Valkov et al., 2016). EXPRESSION / LABELING:
PHENOTYPE:
|
|
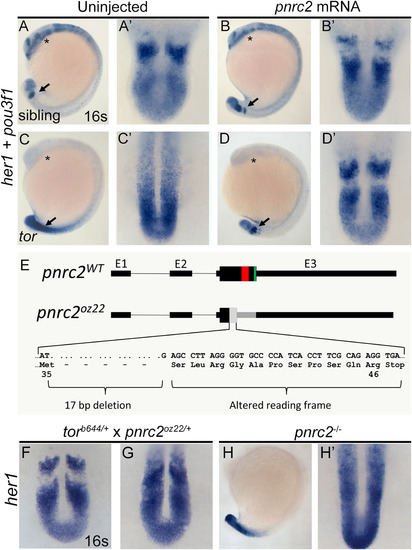
tor mutant embryos are rescued by injection of pnrc2 mRNA and phenocopied by frame-shifting mutation of the pnrc2 locus. Injection of 100 pg pnrc2 mRNA has no effect in wild-type sibling embryos (n=15/15 non-mutant siblings) (A, A’ vs B, B’) but restores striped her1 expression in tor mutant embryos (n=10/10 mutants) (C, C’ vs D, D’; Table 2). In these experiments, rescued torb644 mutant embryos were distinguished from wild-type siblings by lack of expression of pou3f1, a gene located in the torb644 deficiency interval. Asterisks (*) mark the neural pou3f1 expression domain. Arrows mark the her1 expression domain (A-D), magnified in dorsal view to the right of each embryo (A’-D’). Using CRISPR-based mutagenesis, we induced a 17 bp deletion within the pnrc2 coding sequence, creating an early frameshift allele, designated pnrc2oz22 (E). Predicted mutant protein sequence is based on sequenced genomic DNA (E). The pnrc2oz22 allele fails to complement the her1 accumulation phenotype of the torb644 deletion allele (n=4/15 embryos from a heterozygote intercross with tor-like her1 accumulation) (F, G). Similar to torb644 mutants, pnrc2oz22 mutant embryos lack a striped her1 expression pattern (H, H'). EXPRESSION / LABELING:
PHENOTYPE:
|
|
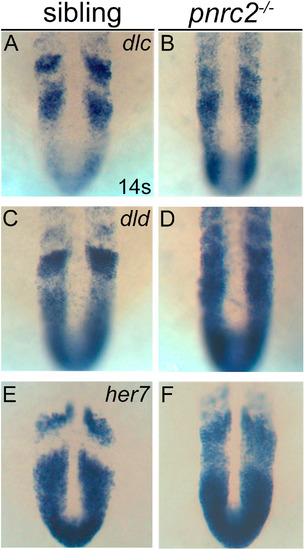
Segmentation clock transcripts accumulate in pnrc2 mutant embryos. Like her1, other segmentation clock genes, dlc (A, B), dld (C,D), and her7 (E, F), are misexpressed in pnrc2 mutant embryos, with expression detected throughout the presomitic mesoderm (PSM) in the expected one-quarter of embryos in a pnrc2oz22intercross, n=8/25 (Χ2=0.65, p=0.4), 6/25 (Χ2=0.013, p=0.9), and 8/25 (Χ2=0.65, p=0.4), respectively (A-F). EXPRESSION / LABELING:
PHENOTYPE:
|
|
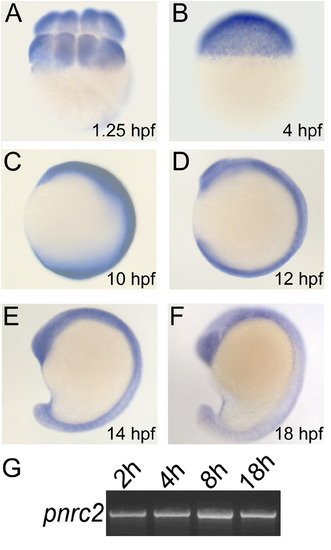
pnrc2 is broadly expressed during segmentation stages. pnrc2 transcripts are detected during early embryonic stages and throughout segmentation stages by in situ hybridization (n>15 per time point) (A-F). As expected, there is no detectable staining with a pnrc2 sense probe (n=15/15) (data not shown). RT-PCR expression analysis for pnrc2 at 2 hours post fertilization (2 h) through mid-segmentation (18 h) is consistent with in situ detection of pnrc2 transcript (see Methods and Results) (G). EXPRESSION / LABELING:
|
|
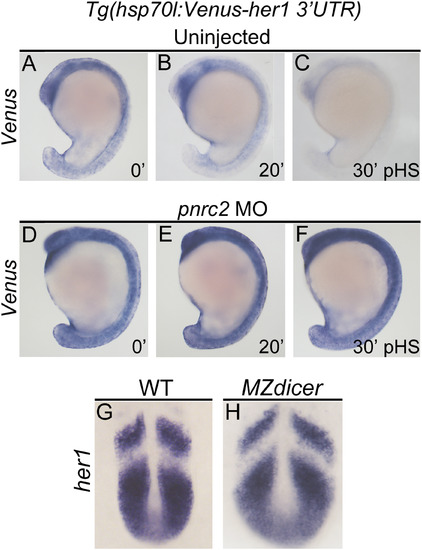
Pnrc2-mediated decay functions via 3’UTR recognition and does not require Dicer-dependent miRNAs. Stably transgenic embryos carrying the hsp70l:Venus her1 3’UTR reporter were injected at the 1-cell stage with pnrc2 sbMO or reserved as uninjected controls. Embryos were then raised to mid-segmentation stage, heat-shocked for 15 min, then collected at the indicated minutes post-heat-shock (pHS) and processed by Venus in situ hybridization (n=65, 6 ng pnrc2 sbMO; n=55, uninjected controls) (A-F). Venus transcripts are not detected in the absence of heat-shock (n=10) (not shown). To determine whether Dicer-generated miRNAs contribute to her1 mRNA decay, MZdicer mutants (n=11) and wild-type controls (n=10) were raised to mid-segmentation stages (16–18 hpf) and processed by her1 in situ hybridization (G, H). At this timepoint, MZdicer mutants are developmentally delayed relative to wild-type controls (Giraldez et al., 2005), and thus have a different overall shape. EXPRESSION / LABELING:
PHENOTYPE:
|
|
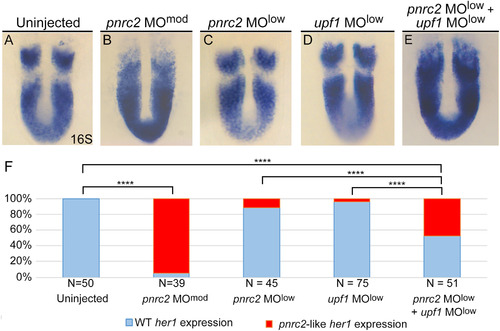
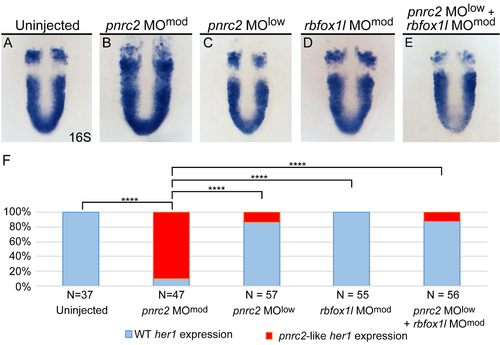
pnrc2 and the nonsense-mediated decay effector Upf1 promote decay of cyclic mRNA. Injection of low dose pnrc2 sbMO (2 ng) has little to no effect on her1 expression (A, C), contrasting with the expected her1 misexpression observed after injection of moderate dose pnrc2 sbMO (6 ng) (B). Low dose injection of upf1 sbMO (0.25 ng) also has little effect on her1 expression (D), but when combined with a low dose of pnrc2 sbMO (2 ng), her1 misexpression is observed in about 50% of injected embryos (E). The proportion affected in each condition is plotted on a bar graph that indicates significant differences between single morphants, double morphants, and controls (F). sbMO = splice-blocking MO; **** = p<0.0001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
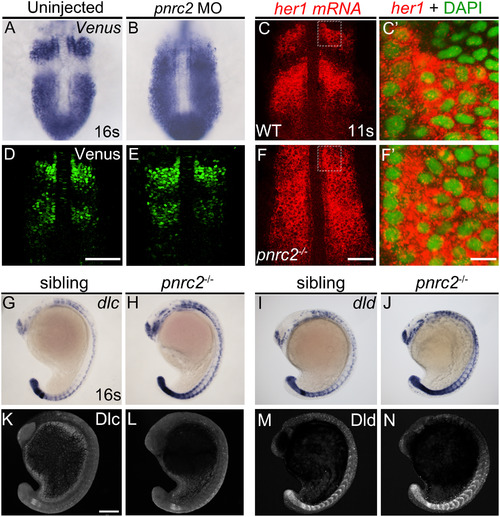
Both reporter and endogenous cyclic transcripts accumulate in Pnrc2-depleted embryos, but protein expression appears normal. Embryos carrying the her1:her1-Venusbk15transgenic clock reporter were injected with pnrc2 splice-blocking morpholino (sbMO) and processed to detect Venus transcripts (A, B) and Venus protein (D, E) at mid-segmentation stages. Representative embryos are shown in A (n=21/21); B (n=18/18), D (n=32/32), and E (n=29/29). Venus immunofluorescence panels (D, E) are at slightly higher magnification than Venus in situ panels (A, B). Detection of her1 mRNA by in situ hybridization chain reaction (HCR-ISH) (C, F) is consistent with chromogenic NBT/BCIP-based in situ detection of endogenous her1 transcript in wild-type and pnrc2 mutant embryos (Fig. 2F, H’), with substantial cytoplasmic localization revealed by DAPI counter staining in 500X magnified view (C’, F’). Because relative intensity of her1 HCR-ISH in pnrc2 mutants to wild-type embryos is high, levels have been reduced in pnrc2 mutant panels (F-F’; see Fig S6 A’’’, B’’’). Misexpression of dlc and dld mRNA is detected throughout the presomitic mesoderm (PSM), formed somites and neurons in the expected one-quarter of embryos in a pnrc2oz22intercross, n=5/28 (Χ2=0.76, p=0.4) and n=7/40 (Χ2=1.2, p=0.3), respectively (G-J). In contrast, Dlc and Dld protein expression is indistinguishable among siblings of the same pnrc2oz22 heterozygote intercross (K-N). Dlc and Dld immunolabeled embryos were genotyped prior to imaging and a subset of wild-type and mutant siblings were imaged by confocal microscopy with representative embryos shown (K-N). Total genotyped individuals per representative panel: n=5 (K), n=6 (L), n=12 (M), n=4 (N). Scale bars=50 µm (D, F), 50 nm (F’), 100 µm (K). EXPRESSION / LABELING:
PHENOTYPE:
|
|
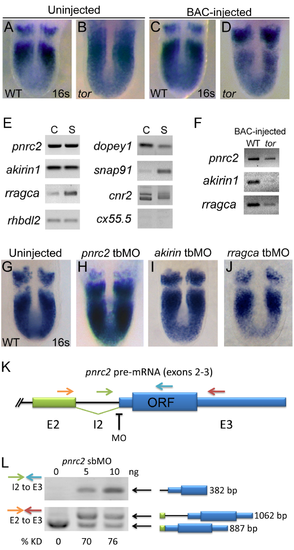
pnrc2 is the best candidate within the tortuga interval for the gene regulating cyclic transcript decay. Injection of 30 pg BAC AL844887 does not affect her1 expression in wild-type sibling embryos (n=27/27) (A versus C), but does significantly reduce accumulation of her1 transcripts in tortugab644 mutants (n=10/10) (B versus D; Table S1). BAC-injected tor mutants are indistinguishable from wild-type siblings and are identified by PCR-based genotyping of individual embryos after her1 in situ hybridization (data not shown). (E) RT-PCR expression analysis of genes on BAC AL844887 reveals that seven are expressed during cleavage (C) and segmentation stages (S), with particularly robust detection of pnrc2 and akirin1. In addition to RefSeq-annotated genes on BAC AL844887, snap91 and dopey1, both of which lack RefSeq annotation, are detectably expressed during cleavage and segmentation stages (E). (F) RT-PCR expression analysis of genes on BAC AL844887 in tortuga mutants indicates that pnrc2, rragca, and to a lesser extent akirin1 are expressed from injected BAC DNA (see Supplemental Methods). dopey1, snap91, cnr2, and rhbdl2 are not expressed from the BAC (data not shown). (G-I) Injection of translation-blocking morpholinos (tbMOs) targeting akirin1 and rragca into 1-cell stage wild-type embryos has no effect on the striped her1 expression pattern observed in WT embryos (G vs I, J), but injection of a pnrc2 tbMO has a dramatic effect (G vs H). A range of doses (1.5 ng-6 ng) was tested for each MO (n>50 per dose). (J) Schematic depicts primer and splice-blocking MO (sbMO) target location on the pnrc2 transcript. (K) RT-PCR splicing analysis was performed to detect both spliced and unspliced pnrc2 transcript in uninjected wild-type and pnrc2 morphant embryos (see Supplemental Methods). One primer combination was used to specifically amplify unspliced product (green and blue arrows in K) and exon-specific primers (orange and red arrows in K) were used to amplify both spliced and unspliced product for semi-quantitative splicing analysis (Gallagher et al, 2011). Injection of pnrc2 sbMO reduces normal pnrc2 splicing by >70% knockdown in a dose-dependent manner (n=20 per dose) (L). Although intron retention alone does not disrupt pnrc2 coding potential as this lies entirely in exon 3 (K), aberrant transcripts induced by pnrc2 sbMO-injection coincide with elevated her1 mRNA levels (Fig 1D) suggesting that pnrc2 sbMOs are effective. Injection of either pnrc2 splice-blocking (sbMO) or a pnrc2 tbMO have similar, dose-dependent effects on her1 expression when injected at doses ranging from 1.5-6 ng (n>50 per dose) (data not shown). All PCR products were sequenced using primers listed in Table S3. ORF=pnrc2 open reading frame; KD=knockdown. |
|
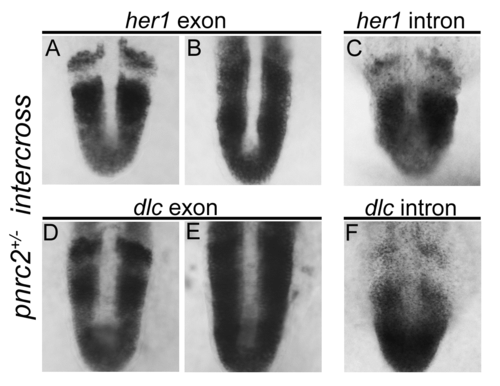
her1 and dlc transcripts accumulate post-transcriptionally in pnrc2oz22mutants. Exonic in situ probes reveal that segmentation clock-associated her1 and dlc transcripts are misexpressed in the expected one-quarter of embryos in a pnrc2oz22 intercross, n=13/57 (X2=0.1, p=0.7) and n=8/30 (X2=0.04, p=0.83), respectively (A, B and D, E). Intronic in situ probes, however, reveal no differences in expression among embryos from the same clutch, n=30/30 (X2=10.0, p=0.0016) and n=45/45 (X2=15.0, p=0.0001), respectively (C, F). These results are consistent with previous observations in torb644 mutants using intronic and exonic in situ probes that distinguish nascent from processed transcripts (Dill et al, 2005). |
|
her1 expression is normal in embryos injected with a control MO, either alone or in combination with a sub-optimal dose of pnrc2 sbMO. Injection of a sub-optimal dose of pnrc2 sbMO (2 ng) has little to no effect on her1 expression (A, C), contrasting with the expected her1 misexpression observed after injection of a moderate dose of pnrc2 sbMO (6 ng) (B). Injection of a moderate dose of an unrelated sbMO targeting rbfox1l (6 ng) either alone (D) or combined with a sub-optimal dose of pnrc2 sbMO (2 ng) has no effect on her1 expression (E). The proportion affected in each condition is plotted on a bar graph that indicates significant differences between single morphants, double morphants, and controls (F). To confirm effective rbfox1l knockdown, we co-injected rbfox1l and rbfox2 sbMOs (6 ng each) in parallel experiments (neither sbMO produces an overt morphological phenotype when injected alone) and observed the expected paralysis phenotype in all double-injected embryos (n=27/27 paralyzed embryos at 24 hpf) (Gallagher et al, 2011; Berberoglu et al, 2017), confirming that we were using an effective rbfoxl1 sbMO dose in our experiments. sbMO = splice-blocking MO; **** = p<0.0001. |
|
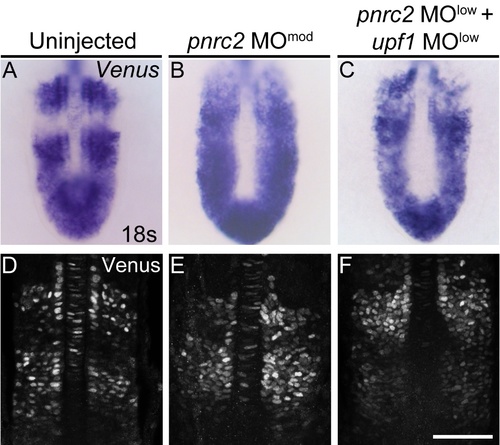
Venus reporter transcripts accumulate in pnrc2- and upf1-deficient embryos, but Venus protein expression appears normal. Embryos carrying the her1:her1-Venusbk15 transgenic clock reporter were injected with a moderate pnrc2 sbMO dose (6 ng) or co-injected with low pnrc2 and upf1 sbMO doses (2 ng and 0.25 ng, respectively) to detect Venus transcripts (A-C) and Venus protein (D-F) at mid-segmentation stages. Representative embryos are shown in A (n=12), B (n=8), C (n=9), D (n=36), E (n=24), and F (n=27). Venus immunofluorescence panels (D-F) are at slightly higher magnification than Venus in situ panels (A-C). sbMO = splice-blocking MO; scale bar = 50 um (D-F). |
|
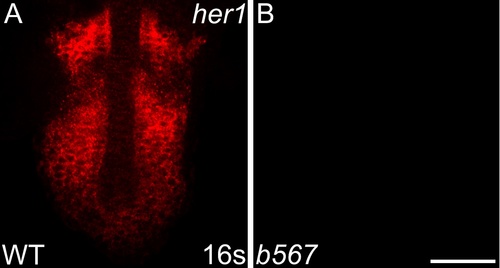
her1 transcripts are not detected in embryos homozygous for a her1 deficiency. Detection of her1 mRNA by in situ hybridization chain reaction (HCR-ISH) by confocal microscopy reveals striped her1 expression in the presomitic mesoderm (PSM) of wild-type embryos (A), mirroring what is observed by colorimetric in situ. As expected, Df(Chr05:her1,her7,ndrg3a)b567 homozygote embryos, imaged using the same laser intensity and exposure as wild-type embryos in A, lack detectable her1 expression (B). Scale bar = 50 um. |
|
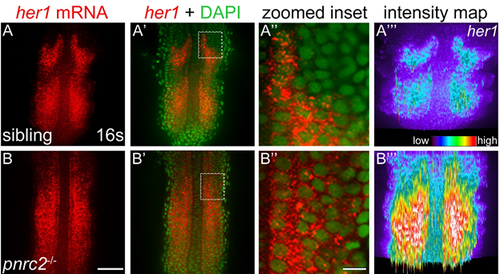
her1 mRNA accumulates dramatically in pnrc2oz22 mutants. Detection of her1 mRNA by in situ hybridization chain reaction (HCR-ISH) reveals her1 misexpression throughout the presomitic mesoderm (PSM) of pnrc2 mutants (A, B). Representative embryos are shown (n=5 in A, n=5 in B). Substantial cytoplasmic localization revealed with DAPI counter staining is apparent in both unaffected siblings and pnrc2 mutants (A', B', 470X magnified in A”, B”). Because detection of her1 mRNA in pnrc2oz22 mutants is very high relative to wild type, levels have been reduced in pnrc2 mutant panels (B-B”). When wild-type and pnrc2 mutants are imaged at the same laser level, raw intensity maps of her1 HCR-ISH signal reveal the extent of her1 mRNA accumulation in pnrc2oz22 mutants (A''', B'''). Scale bars = 50 um (B), 50 nm (B''). |
Reprinted from Developmental Biology, 429(1), Gallagher, T.L., Tietz, K.T., Morrow, Z.T., McCammon, J.M., Goldrich, M.L., Derr, N.L., Amacher, S.L., Pnrc2 regulates 3'UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation, 225-239, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.