- Title
-
A gene trap transposon eliminates haematopoietic expression of zebrafish Gfi1aa, but does not interfere with haematopoiesis
- Authors
- Thambyrajah, R., Ucanok, D., Jalali, M., Hough, Y., Wilkinson, R.N., McMahon, K., Moore, C., Gering, M.
- Source
- Full text @ Dev. Biol.
|
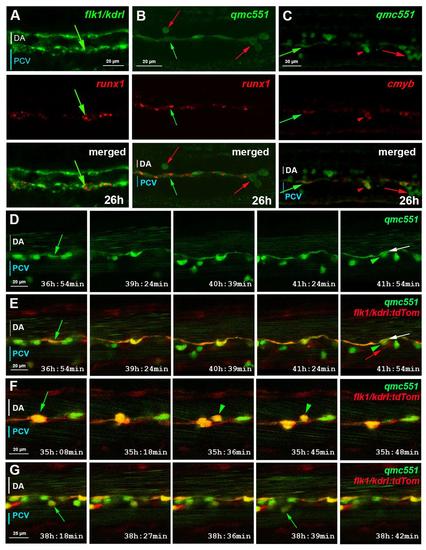
The zebrafish gene trap line qmc551 expresses GFP in primitive red blood cells and in haemogenic endothelial cells of the ventral wall of the dorsal aorta. (A) Structure of the gene trap transposon and strategy of the gene trap screen. (B) Lateral view of a fixed qmc551 embryo. (C) Close-up of the trunk. (D-F) GFP immunohistochemistry and diaminobenzidine staining on a qmc551 embryo. (E) A magnified image of the trunk. (F) A 10 µm transverse section through the trunk of the embryo after plastic embedding. (G) A maximum intensity projection of a 67.5 µm thick confocal Z-stack showing a lateral view of the trunk of a fixed qmc551 embryo. (H) A 1.1 µm YZ cross section of the Z-stack shown in (G). (I,J) Confocal images of the trunk of a qmc551;gata1:dsRed (I) and qmc551;flk1:tdTom (J) double transgenic embryo after fluorescent immunostaining. (K) Confocal images of a live qmc551;csl:cer double transgenic embryo. The csl:cer transgene is a derivative of the csl:venus transgene which we have previously shown to be expressed in arterial ECs ( Gray et al., 2013). (L) Double fluorescent runx1 and flk1/kdrl WISH. (M,N) Fluorescent runx1 (M) and cmyb (N) WISH combined with GFP immunohistochemistry. (O-R) Confocal timelapse microscopy of the DA of qmc551;flk1:tdTom embryos. Images were taken every 3 min. Times on panels represent hours and minutes after fertilization. Note that prRBCs in circulation appear as short lines in the confocal image, while stationary cells are round. The confocal analyses in (I-R) were performed at single cell resolution on 2 (I), 1.8 (J), 2.5 (K), 2.0 (L,N), 1.0 (M) and 2.1 (O-R) µm optical slices. All images (B-E,G,I-R) show embryos with anterior to the left and dorsal up. Red arrows – prRBCs; red arrowheads –- prRBC progenitors trapped in the mesenchyme; green arrows – HECs in the vDA. |
|
qmc551:GFP expression recapitulates gfi1aa expression and marks haematopoietic stem and progenitor cells throughout ontogeny. Views of embryos are posterior in (B) and lateral in (A,C-E,G-I,O). Anteroposterior and dorsoventral axes are indicated. (A-H) Images of live qmc551 embryos and of fixed non-transgenic and qmc551 transgenic embryos after WISH with probes against endogenous gfi1aa and gfp mRNA. (F) Confocal timelapse images (2.0 µm thick optical slice) through the CHT of a 48 hpf qmc551;flk1:tdTom embryo. (I) Images of a qmc551 transgenic embryo immunostained for GFP expression using diaminobenzidine. (J-N). Transverse 10 µm sections of the same embryo after plastic-embedding. The positions along the anteroposterior axis are indicated in (I). (O) Timelapse microscopy of the head region. (P) GFP immunohistochemistry and diaminobenzidine staining on 10 µm sections of adult kidneys isolated from wt, qmc551 and cd41:gfp fish. (Q) Flow cytometric analysis of KM cells of wt and qmc551;gata1:dsRed double transgenic adults. GFP/dsRed fluorescence and forward/side scatter were analyzed. Excitation and detection wavelengths are indicated in nm. Cell populations were gated according to Traver et al. (2003). (R-U) Cytocentrifugation and Giemsa staining of the qmc551:GFP+ KM cells identified neutrophils (R), macrophages (S), progenitors (T) and lymphocytes (U). Annotations: caudal haematopoietic tissue (CHT), glomerulus (Gl), goblet cell (GC), gut (G), inner ear (IE), lateral line organ (LL), medial crista (MC), mesenchyme (M), notochord (Nc), pancreas (P), posterior blood island (PBI), putative Paneth cell (PC), pharyngeal sensory cells (asterisks), posterior macula (PM), swim bladder (SB), somitic muscle (SM), thymus (T) and ventral wall of the DA (vDA) and of the caudal artery (vCA). HSPCs, pRBCs, thymocytes and wandering leukocytes are labeled with green, red, white and turquoise arrows, respectively. EXPRESSION / LABELING:
|
|
Gfi1aa expression in haemogenic endothelial cells is induced downstream of VegfA and Notch signaling, but independent of Runx1. Fixed qmc551;gata1:dsRed double transgenic embryos after GFP/dsRed immunohistochemistry are shown in (A, D-E, I). Fixed wt, qmc551 and qmc551;gata1:dsRed double transgenic embryos stained by WISH are shown in (B-C, H, K), (L) and (J), respectively. Live qmc551 embryos that were wt, heterozygous or homozygous mib carriers were imaged in (F-G). Confocal images of optical sagittal sections through the DA are 1.6 and 0.995 µm (A), 6.5 and 6.6 µm (D), 1.2 µm (E) and 2.7 µm (I) thick. A confocal maximum intensity projection of a 37 µm optical slice is shown in (G). Embryos were treated with DMSO, the VegfR inhibitors 676,475 (A) and SU5416 (B,C) or DAPM (D) from tailbud stage (10 hpf). Rbpja/b (E) and runx1 (I-L) morpholinos were injected at 2–4 cell stage. PrRBCs, HECs and inner ear hair cells are labeled with red, green and yellow arrows, respectively. Arrowheads mark reduced or absent staining. Fractions x/y give the number of embryos, x, with depicted phenotype out of all embryos analyzed, y. Embryos are shown with anterior left and dorsal up. |
|
Gfi1aa expression is lost in primitive erythrocytes of homozygous qmc551 embryos, yet primitive haematopoiesis is unaffected. (A) Genomic organization of the gfi1aaqmc551Gt locus. Oligos I and J were used in the RT-PCR experiments. The sequence complementary to the gfi1aa WISH probe is indicated. The TaqMan probe overlapped with the exon 1/2 boundary and is not shown. (B) Fluorescent images of 20 hpf wt, qmc551 heterozygous and homozygous siblings from an incross of qmc551 heterozygous carriers. (C,H,I,L,M) RNA WISH experiments with indicated probes on 13-20 hpf wt and qmc551 homozygous embryos. Views are posterior with anterior up on all images of 13-16 hpf and lateral with anterior to the left and dorsal up on all images of 18, 20 and 22 hpf embryos. (D) QRT-PCR to determine relative levels of gfi1aa mRNA in 16 hpf whole embryos. EF1α mRNA was used as a loading control. (E) Flow cytometric analysis of embryonic cells at 19 hpf showing green and red fluorescence excited with a 488 nm laser, and detected using the band pass filters 529/28 and 580/23 (central wavelength/width in nm), respectively. (F) Mean green fluorescence observed in GFP+ cells in embryos at 19 hpf. Please note that values are shown relative to heterozygous controls (Two-tailed t-test: p<0.0001). (G) The relative numbers of GFP+ cells in embryos at 19 hpf. (J,Q) Diaminofluorene staining to detect hemoglobin in 3 dpf embryos. (K,R) Images of prRBCs that were isolated from the sinus venosus of terminally anaesthetized 3 day-old embryos and stained with May-Grünwald and Giemsa. (N) Structure of the gfi1b transcript before and after splicing in the presence and absence of gfi1b morpholinos. In the morphant, exon 3 and 5 sequences are spliced together (dashed line). (O) RT-PCR performed on RNA isolated from uninjected and morpholino-injected wt embryos. (P)gfi1b cDNA sequence representing the alternatively spliced gfi1b mRNA isolated from gfi1b morphant embryos. PCR oligo and divergent C-terminal amino acid sequences are underlined. Sequences derived from different exons are shown in different colors. The numbering of nucleotides corresponds to that of database entry NM_001271841. Encoded amino acids are counted below. Arrows: red - prRBCs, yellow - inner ear hair cells, light blue - anterior lateral mesoderm, orange - hatching gland precursors. Arrowheads: red - prRBCs in heart. Fractions x/y give the number of embryos, x, with depicted phenotype out of all embryos analyzed, y. Embryos are shown with anterior left and dorsal up. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Gfi1aa expression is lost in definitive haematopoietic cells of homozygous qmc551 fish, but definitive haematopoiesis is normal. (A,C,F-O) Fixed embryos stained in WISH experiments using indicated probes. Whole embryos are shown in (A,H,J,M-O). Close-up views of anterior and posterior parts of the embryos are presented in (G) and (C,F,I,K), respectively. All embryos, except (G), are shown in a lateral view with anterior to the left and dorsal up. (G) shows a close-up dorsal view with anterior to the left. (L) shows a transverse section through the trunk of one of the homozygous qmc551 embryos after gfi1ab WISH. (B,R) QRT-PCR on total RNA isolated from whole 26 hpf embryos (B) and from adult KM cells (R) to measure the relative level of gfi1aa mRNA (Two-tailed t-test: p=0.002 (26 h), p=0.034 (KM)). Ef1α mRNA was used as a loading control. (D,E) Live images of qmc551 embryos with anterior left and dorsal up. Close-up pictures of the tail (D) and the head regions (E) are presented. (P) Regulation of Gfi1aa and Gfi1ab expression at the onset of definitive haematopoiesis. (Q) Flow cytometric analysis of green and red fluorescence in adult KM cells excited with a 488 nm laser and detected using 529/28 and 580/23 nm band pass filters, respectively. Please note that a substantial proportion of KM cells displays green and red autofluorescence. (S) May-Grünwald/Giemsa stained KM cytospins. (T) Relative number of bilobed and trilobed neutrophil granulocytes observed in May-Grünwald/Giemsa stained cytospins. (U) Sudan Black staining on adult KM cytospins. Arrows: red - prRBCs, green - HECs and definitive HCs, yellow - inner ear hair cells, white - T cell precursors in the thymus, black - neutrophil granulocytes with bilobed nuclei. Arrowheads: green - reduced gene expression in the vDA. EXPRESSION / LABELING:
PHENOTYPE:
|
|
In the gene trap line qmc551, GFP is expressed in primitive erythrocytes and in endothelial cells of the ventral wall of the dorsal aorta. This figure supports data shown in Fig. 1. (A) Single color and merged confocal images of the trunk of a qmc551;gata1:dsRed double transgenic embryo after fluorescent GFP and dsRed immunostaining. (B) Single color and merged confocal images of a qmc551;flk1:tdTom double transgenic embryo after fluorescent immunostaining for GFP and tdTom. (C) Single color and merged confocal images of a live qmc551;csl:cer double transgenic embryo. Note that prRBCs in circulation appear as short lines in the confocal image in (C), while stationary cells are round. Images were taken at single cell resolution on 2 (A), 1.8 (B) and 2.5 (C) µm optical slices. Times given on the panels represent hours and minutes after fertilization. All images show embryos with anterior to the left and dorsal up. Red arrows - prRBCs; red arrowheads - prRBC progenitors trapped in the mesenchyme; green arrows - ECs in the vDA. |
|
qmc551:GFP positive endothelial cells are haemogenic endothelial cells. This supplementary figure is related to Fig. 1. Confocal images of fixed (A-C) and live (D-G) embryos show optical sagittal sections through the DA with anterior left and dorsal up. The optical slices were at single cell resolution thickness, i.e. 2.0 (A,C), 1.0 (B) and 2.1 µm (D-G). Images in (D-G) are taken from timelapse experiments in which pictures were taken every 3 minutes. Times on panels represent hours and minutes after fertilization. (A) Double fluorescent runx1 and flk1/kdrl whole-mount in situ hybridisation (WISH). (B-C) Fluorescent runx1 (B) and cmyb (C) WISH combined with GFP immunohistochemistry. prRBCs (red arrows); prRBC progenitors in mesenchyme (red arrowheads) and HECs (green arrows). (D-G) Timelapse microscopy on qmc551;flk1:tdTom embryos from 31 hpf. (D) shows GFP expression only, while (E-G) present merged images. Annotations: HECs before (green arrow) and after bEMT (green arrowhead); DA endothelium after bEMT of HEC (white arrow); venous EC (red arrow). |
|
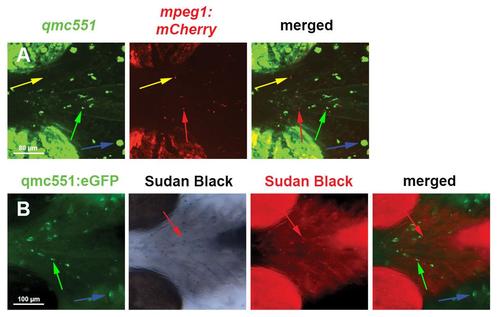
Most GFP-positive cells that patrol the head tissues of qmc551 transgenic embryos do not express markers of mature macrophages and neutrophil granulocytes. (A) Live embryos that carry the qmc551 gene trap and the macrophage reporter transgene mpeg1:mCherry display numerous GFP (green arrow) and mCherry (red arrow) single-positive cells, but hardly any double-positive cells (yellow arrow). (B) Fixed qmc551 transgenic embryos stained for the mature neutrophil marker Sudan Black and immunostained for GFP harbor many cells that are single-positive for GFP (green arrow) and for Sudan Black (black in the original image; red in the pseudocolored image; red arrow). All images show ventral views of the head region of the embryo, with anterior to the left. |
Reprinted from Developmental Biology, 417(1), Thambyrajah, R., Ucanok, D., Jalali, M., Hough, Y., Wilkinson, R.N., McMahon, K., Moore, C., Gering, M., A gene trap transposon eliminates haematopoietic expression of zebrafish Gfi1aa, but does not interfere with haematopoiesis, 25-39, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.