- Title
-
Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development
- Authors
- Ng, A., Uribe, R.A., Yieh, L., Nuckels, R., and Gross, J.M.
- Source
- Full text @ Development
|
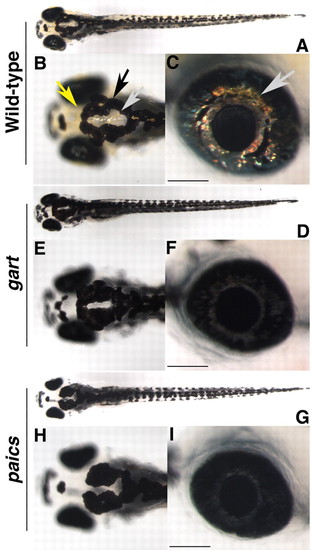
gart and paics mutants have pigmentation defects and microphthalmia. (A-G) Dorsal (A,B,D,E,G,H) and lateral (C,F,I) images of wild-type (A-C), gart (D-F) and paics (G-I) embryos at 5 dpf. Wild-type embryos possess xanthophore (yellow arrow), melanophore (black arrow) and iridophore (gray arrow) derived pigments. gart and paics mutants have substantially less xanthophore and iridophore pigmentation and reduced levels of melanin pigmentation. Mutants are also microphthalmic. Scale bar: 100μm. PHENOTYPE:
|
|
Histological analysis of gart and paics mutants. (A-J) Transverse histological sections of wild type (A,C,E,H), gart (D,G,J) and paics (B,F,I) mutants. (A,B) paics mutants are identifiable at 48 hpf and are microphthalmic with a hypopigmented RPE. Retinal lamination and neuronal differentiation are also delayed. (C,D) gart mutants are morphologically obvious at 54 hpf and are microphthalmic with a hypopigmented RPE. Retinal lamination and neuronal differentiation are also delayed. (E-G) At 72 hpf, mutant eyes remain microphthalmic, however, the retina is well laminated and all cell types appear to be present. (H-J) At 5 dpf, mutant eyes remain microphthalmic. Scale bar: 100 μm. Dorsal is up. |
|
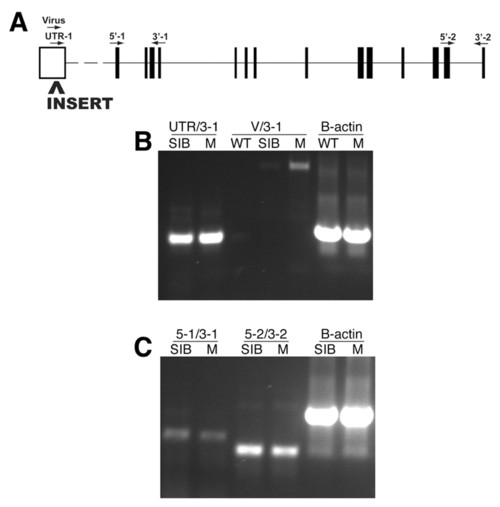
Molecular analysis of the garthi3526b locus. (A) The approximate position of the proviral insertion and RT-PCR primers mapped onto a schematic of the mutated gene (www.ensembl.org/Danio_rerio/). The proviral insertion is located in the predicted 5′ UTR. Amplification of the region between the UTR and exon 4 (B, UTR/3-1), the region between exons 1 and 4 (C, 5-1/3-1), or the region between exons 13 and 14 (C, 5-2/3-2) all indicate that the overall levels of gart transcripts are unaffected in the homozygous mutant embryos. Amplification from the 3′ end of the proviral insert to exon 4 (B, V/3-1) in homozygous gart mutants indicates that a portion of the proviral insert is transcribed and retained in the resulting mRNA. WT, wild-type AB; SIB, wild-type sibling mixture (+/+ and +/-); M, homozygous mutant. |
|
Molecular analysis of the paicshi2688 locus. (A,B) The approximate position of the proviral insertion and RT-PCR primers mapped onto a schematic of the mutated gene (www.ensembl.org/Danio_rerio/). (A) paicsLONG is encoded by at least 15 exons, and all of the conserved enzymatic domains of the Paics protein are found in the sequence encoded by exons 7-15. The proviral insert in paics mutants is located in exon 7 of paicsLONG. (B) paicsSHORT is composed of the latter nine exons of paicsLONG, in addition to a tenth noncoding first exon. The proviral insert in paics mutants is located in exon 2 of paicsSHORT. (C) RT-PCR using primer sets that amplify the region between exons 4 and 9 of paicsLONG (3F/4R), and exons 1 and 4 of paicsSHORT (UTR/4R) indicates that both transcripts are expressed in wild-type embryos. Amplification of the region surrounding the site of proviral insertion (3F/4R) in paicsLONG indicates that the proviral insert leads to an alteration in mRNA splicing in the paicsLONG transcript. (D) RT-PCR using primer sets that assay for the presence of the proviral insert in paicsSHORT (UTR/V) indicates that the viral insert is present in paicsSHORT transcripts. RT-PCR amplification using paicsSHORT-specific exon 2 primers located downstream of the site of proviral insertion (7F/4R and 7-8F/4R) indicates that the virus sequence in paicsSHORT does not lead to detectable changes in transcript levels. WT, wild-type AB; SIB, wild-type sibling mixture (+/+ and +/-); M, homozygous mutant. |
|
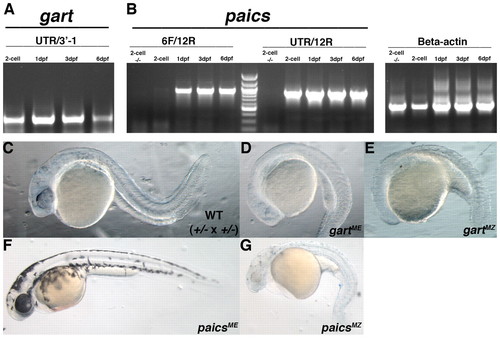
gart and paics are maternally provided and limit the severity of phenotypes in zygotic mutants. (A) RT-PCR analysis of gart indicates maternal expression at the 2-cell stage and sustained zygotic expression at 1, 3 and 6 dpf. (B) RT-PCR analysis of paicsLONG (6F/12R) and paicsSHORT (UTR/12R) at the 2-cell stage and 1, 3 and 6 dpf. Only paicsSHORT is provided maternally, whereas maintained zygotic expression of both splice forms was detected at 1, 3 and 6 dpf. beta actin was used as a control. (C) Wild-type sibling derived from a gart+/- x gart+/- cross at 28 hpf. (D) gart maternal-effect mutants (gartME) at 28 hpf are drastically delayed in development when compared with wild-type embryos derived from heterozygous incrosses. (E) gart maternal-zygotic (gartMZ) mutants at 28 hpf have arrested in development and display severe defects in anterior development. (F) paics maternal-effect mutants (paicsME) at 52 hpf have pigmentation defects and microphthalmia but are relatively normal otherwise. (G) paics maternal-zygotic (paicsMZ) mutant at 52 hpf. paicsMZ mutants are small, nearly albino and possess cardiac edemas and widespread CNS degeneration. |
|
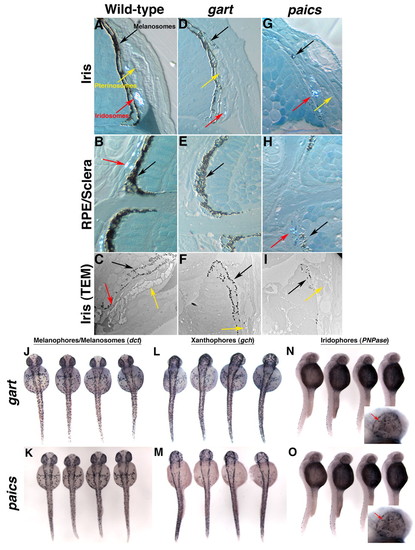
Pigment-containing organelles and cell types are normal in gart and paics mutants. (A-I) High magnification images of transverse histological sections through wild-type (A,B), gart (D,E) and paics (G,H) eyes indicating the presence of melanosomes (black arrow), pterinosomes (yellow arrow) and iridosomes (red arrow) in the pigmented iris (A,D,G) and of melanosomes and iridosomes in the RPE and sclera (B,E,H). (C,F,I) TEM images of the iris of wild type (C), gart (F) and paics (I). (J-O) In situ hybridization for pigment cell types in representative embryos derived from gart (J,L,N) and paics (K,M,O) heterozygous incrosses. (J,K) dct marks melanocytes and melanosomes in the RPE. (L,M) gch marks xanthophores. (N,O) PNPase marks iridophores. Inset in N and O, high magnification view of iridophores in the eye (arrows). No differences in the number or distribution of pigment cells were observed between wild-type and mutant embryos (not shown). |
|
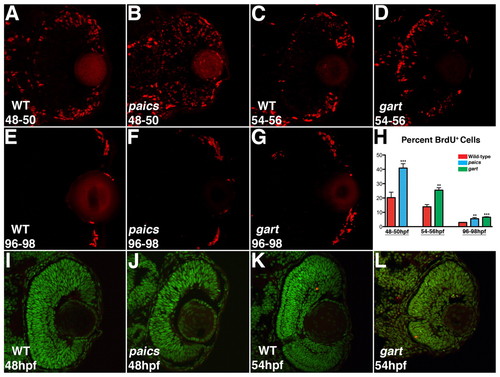
Cell proliferation and apoptosis in gart and paics mutants. (A,C) BrdU exposures 48-50 hpf and 54-56 hpf indicate that in the wild-type eye most proliferative cells are located at the retinal periphery. (B) In paics and (D) gart mutants, BrdU+ cells are located throughout the central retina. (E-G) BrdU exposures 96-98 hpf localized proliferative cells in the wild-type and both paics and gart mutant retinas to the retinal periphery. (H) Quantifying the percentage of BrdU+ cells in each retina identified significant increases in the paics and gart mutants at each time point (** P<0.05, *** P<0.001). (I-L) TUNEL labeling to identify apoptotic nuclei. No increases in apoptosis over wild-type levels were observed in paics mutants at 48 hpf (I,J) or gart mutants at 54 hpf (K,L). |
|
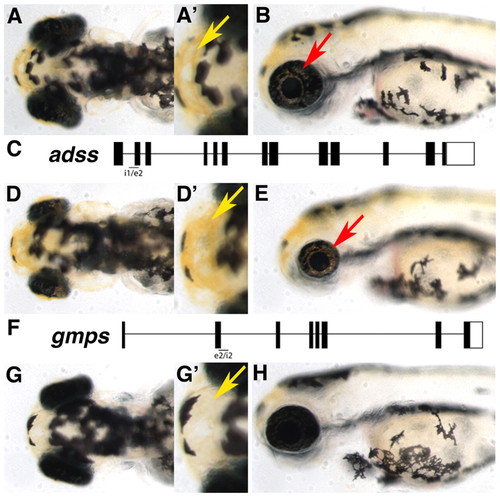
Requirement for adss in eye growth and gmps in pigmentation downstream of gart and paics. (A-B) Dorsal (A,A′) and lateral (B) images of control MO-injected embryos at 3 dpf. Xanthophores (yellow arrow) and iridophores (red arrow) are highlighted. (C) The adss locus and approximate position of the morpholino target site. (D-E) adss morphants are microphthalmic but have normal pigmentation. (F) The gmps locus and approximate position of the morpholino target site. (G-H) gmps morphants have nearly normal sized eyes but lack most xanthophore- and iridophore-derived pigmentation. Xanthophores can be observed (G′) but are markedly lighter than those in control-injected embryos (A′). PHENOTYPE:
|
|
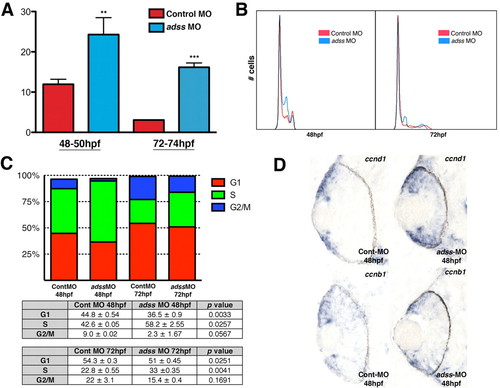
The ATP pathway is required for S-phase progression. (A) BrdU exposures 48-50 hpf and 72-74 hpf indicate a higher proportion of S-phase cells (y-axis) in adss morphants (** P<0.05, *** P<0.005). (B) DNA content analysis plots from flow cytometry at 48 hpf and 72 hpf. (C) Graphical and tabular data for percentage of cells in G1, S or G2/M, as determined by flow cytometry. adss morphants possess elevated percentages of cells in S phase and decreased percentages in G1. (D) ccnd1 expression is expanded into the central retina of adss morphants when compared with control MO-injected embryos. ccnb1 distribution is normal. EXPRESSION / LABELING:
PHENOTYPE:
|
|
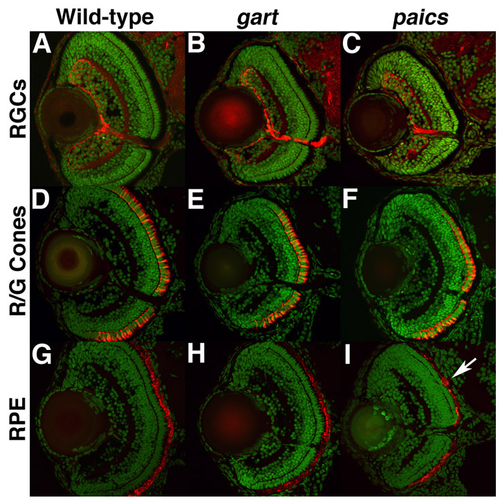
Immunohistochemical analysis of retinal patterning in gart and paics mutants. Transverse retinal cryosections from 5 dpf embryos stained with zn8, a marker of RGCs (A-C); zpr1, a marker of red/green cones (D-F); and zpr2, a marker of the RPE (G-I). No differences between wild type (A,D,G), gart (B,E,H) or paics (C,F,I) were observed, although occasional clumps of RPE were noted in some mutants (arrow in I). |
|
A subset of homozygous gart and paics mutants are viable and develop into normal adults. Heterozygous (A) and homozygous (B) gart mutants at 90 dpf. Heterozygous (C) and homozygous (D) paics mutants at 60 dpf. Both gart and paics homozygous mutants develop into normal adult fish and show no overt adult phenotypes. |
|
adss and gmps morpholino efficacy. (A) RT-PCR for adss using two primer sets that flank the intron 1/exon 2 targeted region of the transcript. Both primer sets do not amplify any transcript from adss-MO-injected embryos at 24 hpf. wnt5 primers are used as a positive control for each cDNA. (B) RT-PCR for gmps using a primer set flanking the exon 2/intron 2 targeted region of the transcript. gmps-MO injection leads to a splicing defect in gmps transcripts at 24 hpf. (C) gmps-MO injection leads to the removal of an 81 bp portion of exon 2 (exon 2 sequence in black, excised portion in red). |

Unillustrated author statements PHENOTYPE:
|