- Title
-
Efficient knock-in method enabling lineage tracing in zebrafish
- Authors
- Mi, J., Andersson, O.
- Source
- Full text @ Life Sci Alliance
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
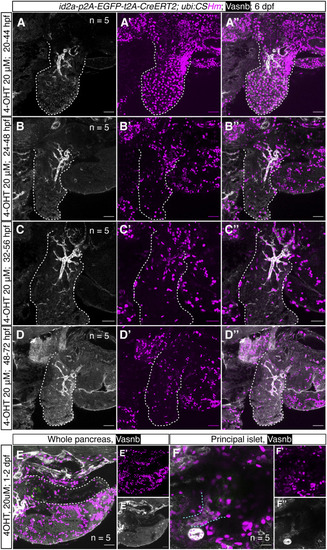
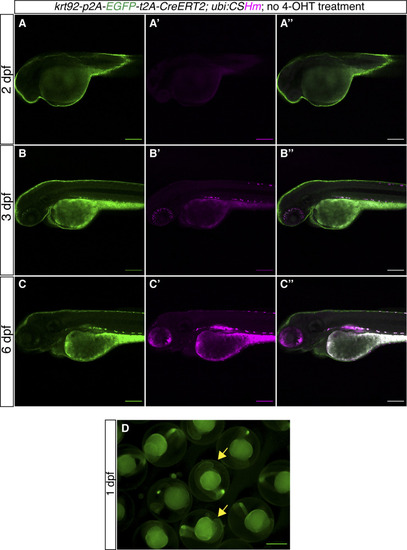
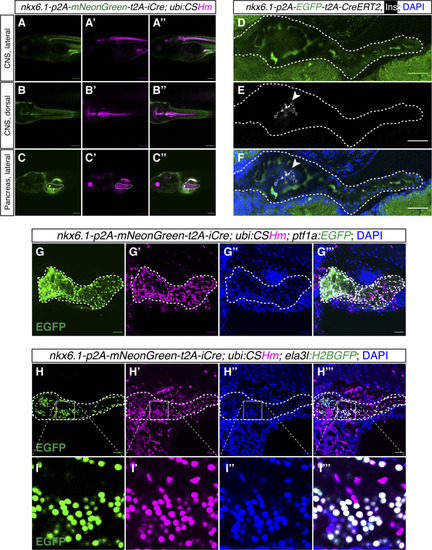
The sequences of different cassettes are shown using different colors. The mutated sequences in the left homologous arms are highlighted with a dark green background color. The introduction of these point mutations keeps the amino acid sequence intact without cleavage by the Cas9/gRNA complex meant to only cleave the genomic insertion site. |
|
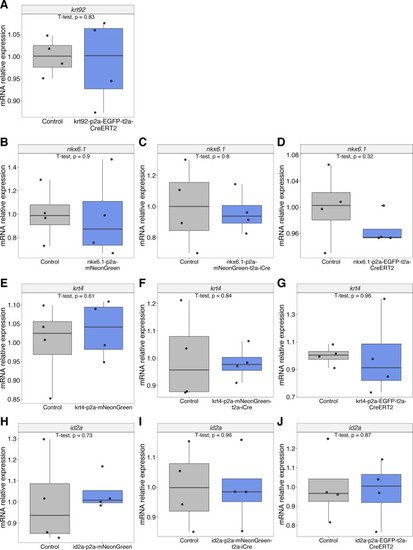
Sanger sequencing of the integrations at the junction of the last exon to the integration for each of the 10 generated knock-in lines. The chromatograms display the sequences, the reverse direction, and show integrations in-frame with the endogenous gene without indels at any of the junctions. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|