- Title
-
Essential Role of Nr2f Nuclear Receptors in Patterning the Vertebrate Upper Jaw
- Authors
- Barske, L., Rataud, P., Behizad, K., Del Rio, L., Cox, S.G., Crump, J.G.
- Source
- Full text @ Dev. Cell
|
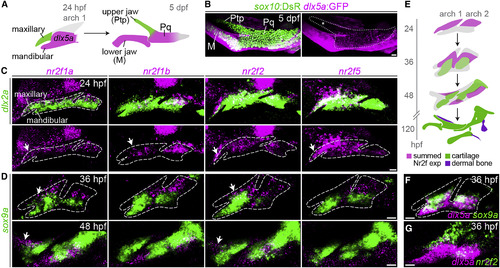
Enrichment of Nr2f Genes in the Maxillary Domain (A) The pterygoid process (Ptp) of the larval upper jaw derives from cells in the maxillary prominence (green), whereas the body of the palatoquadrate cartilage (Pq) derives from a large dlx5a+ pre-cartilaginous condensation (magenta) that also gives rise to the lower-jaw Meckel's cartilage (M). (B) dlx5a:GFP staining (magenta) in the body of the Pq but not the Ptp (asterisk) confirms their distinct origins. Chondrocytes are labeled by sox10:DsRed (green). The dotted line demarcates the full Ptp-Pq element. (C) Double-fluorescence in situ hybridizations for four Nr2f genes (magenta) at 24 hpf, with dlx2a (green) marking arch NCCs. Images are representative maximum-intensity projections. Also see Figure S1 for quantitative expression data and Table S3 for in situ probe information. (D) Nr2f expression (magenta) is largely excluded from nascent sox9a+ pre-chondrocytes (green) at 36 and 48 hpf. Top images are single z slices; bottom images are maximum-intensity projections. (E) Summed Nr2f expression domains in the first and second arches and relative to developing cartilage elements. (F) dlx5a co-localizes with sox9a expression (single z slice). (G) Co-staining shows a sharp boundary between maxillary nr2f2 and mandibular dlx5a expression (maximum-intensity projection). White arrows in (C and D) indicate maxillary expression. Dashed lines in (C, D, and F) reflect approximate arch boundaries. Scale bars, 20 μm. |
|
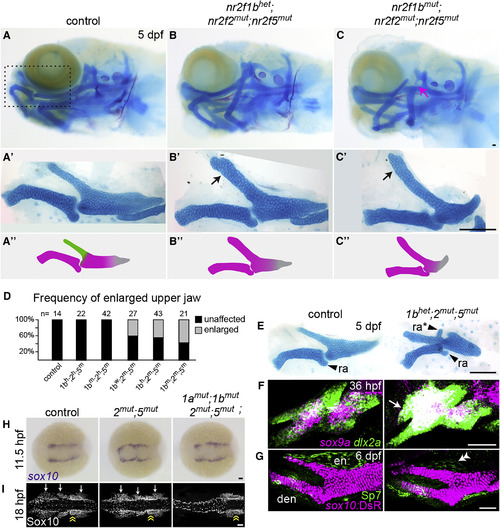
Transformation of the Upper Jaw in Nr2f Mutants (A–C) Larval heads stained for cartilage (Alcian blue) and bone (Alizarin red). Boxed area in (A) represents the approximate region of the dissected upper- and lower-jaw cartilages in A′–C′. In mutants (B′–C′), the pterygoid process (Ptp, green in A″) of the palatoquadrate is thickened (black arrows) to resemble Meckel's (magenta in A″). Skeletal structures derived from the dorsal hyoid arch are reduced in the triple mutant (magenta arrow in C). (D) Frequency of the thickened Ptp phenotype across different nr2f1b; nr2f2; nr2f5 genotypes. (E) Some nr2f2; nr2f5 mutants form an ectopic process (ra∗) that resembles the retroarticular process (ra) of Meckel's. (F) Fluorescent in situ images show expansion of sox9a expression into the mutant maxillary prominence (white arrow; dlx2a labels all arch NCCs). (G) Formation of a larger cartilage (sox10:DsRed+, magenta) in the mutant is accompanied by a reduction in the number of Sp7+ osteoblasts (green) in the neighboring dermal entopterygoid bone (en; white double arrowhead). Mutants have normal numbers of Sp7+ osteoblasts in the lower-jaw dentary bone (den). (H) sox10+ NCCs are specified at the neural plate border in all Nr2f mutants (n = 18 nr2f1aany; nr2f1bany; nr2f2−/−; nr2f5−/−; n = 2 quadruple). The nr2f2; nr2f5 double mutant shown is also nr2f1a+−-; nr2f1b−/−. (I) Three streams of Sox10+ NCCs (white arrows) are evident in all mutant combinations (n = 13 nr2f1aany; nr2f1bany; nr2f2−/−; nr2f5−/−) except the quadruple mutant (n = 1). Sox10+ otic cells (yellow double arrow) are indicated for reference. Scale bars: (C and E) 100 μm; (F–I) 50 μm. Also see Figure S2 and Table S4 for mutation details and skeletal phenotypes of single and other combinatorial mutants. EXPRESSION / LABELING:
PHENOTYPE:
|
|
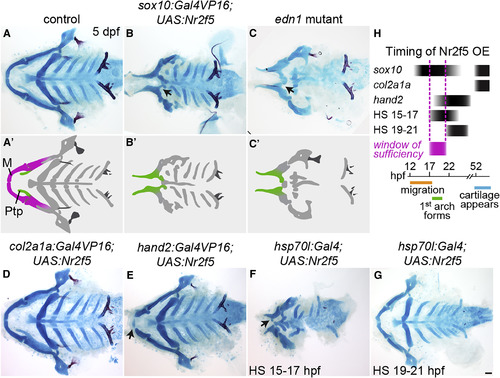
nr2f5 Misexpression Reduces and Transforms the Lower Jaw (A–G) Alcian blue and alizarin red staining of dissected facial skeletons shown from the ventral perspective. The upper jaw (Ptp, green in A′–C′) remains, but the lower jaw (M, magenta in A′) is reduced and transformed (black arrows) in sox10:Gal4VP16; UAS:Nr2f5el662 transgenic fish (B), similar to edn1 mutants (C). Putative transformations are schematized in (A′–C′). Late-onset nr2f5 misexpression in chondrocytes (col2a1a:Gal4VP16) did not affect the facial skeleton (D). Misexpression in post-migratory mandibular NCCs (hand2:Gal4VP16) reduced the distal tips of Meckel's (black arrow, E). Heat-shock-induced embryo-wide misexpression (hsp70l:Gal4) reduced the lower jaw (black arrow) when applied at 15–17 (F) but not 19–21 hpf (G). (H) Approximate timing of Nr2f5 protein overexpression (OE) in the different driver lines, with the approximate window of sufficiency indicated in magenta. Scale bar, 50 μm. Also see Figure S3 for additional analyses of the sox10:Gal4VP16; UAS:Nr2f5 phenotype. PHENOTYPE:
|
|
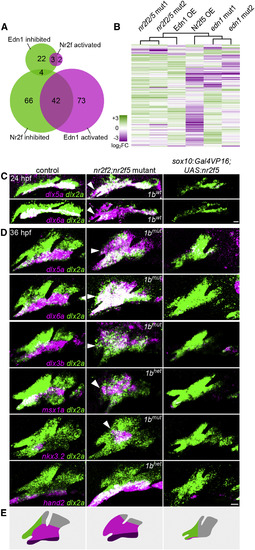
Nr2f2/5 Broadly Inhibit Mandibular Gene Expression (A) Venn diagrams depicting the overlap between genes inhibited and activated by Nr2fs and Edn1. (B) Hierarchical clustering of log2 fold-change values relative to controls for 350 arch-enriched genes in nr2f2; nr2f5 mutants, sox10:Gal4VP16; UAS:Nr2f5 transgenics (Nr2f5 OE), edn1 mutants, and hsp70l:Gal4; UAS:Edn1 (Edn1 OE) transgenics. Reference data are from Askary et al. (2017). See Table S1 for transcripts per million (TPM) values of all arch-enriched genes. (C and D) Fluorescence in situ images for dysregulated genes (magenta) relative to all arch NCCs (dlx2a+, green) in nr2f2; nr2f5 mutants (nr2f1b genotype indicated) and sox10:Gal4VP16; UAS:Nr2f5 transgenics at 24 (C) or 36 hpf (D). Arrowheads indicate ectopic maxillary expression. Images are representative maximum-intensity projections. Scale bars, 20 μm. (E) Schematized shifts in arch gene expression territories. See Figure S4 for expression patterns of non-mandibular genes. |
|
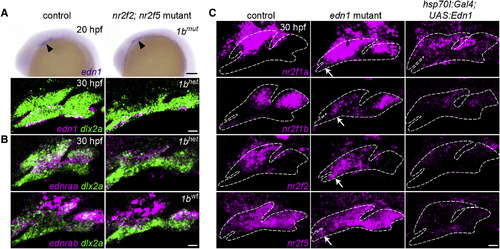
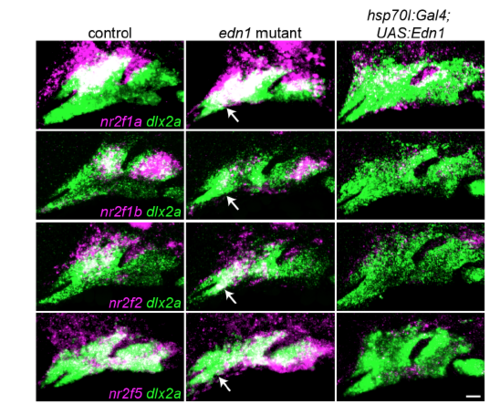
Edn1 Inhibits Nr2f Expression in the Mandibular Prominence (A) edn1 is expressed comparably in the ventral ectoderm of wild-type controls and mutants at 20 hpf (arrowheads) and 30 hpf (magenta). (B) Expression of the Edn1 receptors is reduced (ednraa) or largely unaffected (ednrab) in Nr2f mutants. Arch NCCs are labeled with dlx2a (green), and nr2f1b genotypes are indicated. (C) In edn1 mutants, expression of all four Nr2f genes (magenta) ectopically increases to varying degrees in the mandibular domain (white arrows). Misexpression of edn1 induced by a heat shockof hsp70l:Gal4; UAS:Edn1 embryos from 20 to 24 hpf reduced Nr2f expression throughout the arches. Dashed lines reflect arch boundaries. Images are maximum-intensity projections. Merged images with dlx2a arch NCC staining are presented in Figure S5. Scale bars, (A, top) 100 μm, (A, bottom, B, C) 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
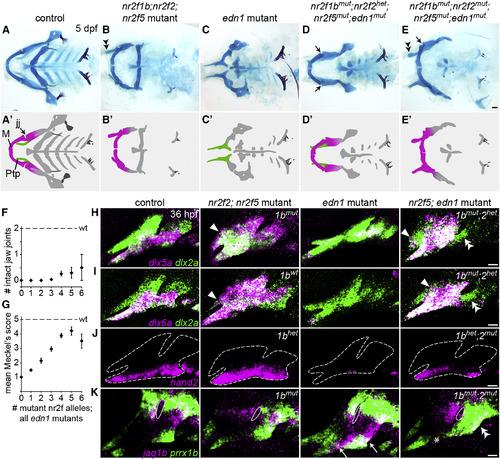
Reducing Nr2f Gene Dosage Can Fully Rescue the Lower Jaw in edn1 Mutants (A–E) Facial skeletons of fish carrying nr2f and/or edn1 mutations, schematized in (A′–E′). Compared to the control (A), nr2f2; nr2f5 mutants (B) have a thickened upper jaw (double arrowhead), while edn1 mutants (C) have a much reduced lower jaw. Reducing Nr2f alleles in edn1 mutants (D and E) can rescue the jaw joint (black arrows) and Meckel's cartilage (compare C with D–E). The upper jaw is still enlarged in nr2f2; nr2f5; edn1 combinatorial mutants (E, double arrowhead). M, Meckel's cartilage; Ptp, pterygoid process; jj, jaw joint. Scale bar in (E) 50 μm. (F and G) Quantification of jaw joint (F) and Meckel's rescue (G) across different summed nr2f; edn1 genotypes. One or both jaw joints were rescued in 12/106 mutants lacking three to six Nr2f alleles. See Figure S6 for Meckel's scoring system. n = 16, 29, 30, 53, 42, 10, and 2 for edn1 mutants with zero to six mutant Nr2f alleles, respectively. Error bars are the SEM. Scores in (G) were non-evenly distributed across genotype classes (p < 0.0001, chi-squared). (H–J) dlx5a (H), dlx6a (I), and hand2 (J) (magenta) are expanded in nr2f2; nr2f5 mutants (white arrowheads), reduced in edn1 mutants, and restored in the first arch of nr2f5; edn1 mutants (nr2f1b/nr2f2 genotypes indicated). Some ectopic expansion into maxillary cells was still evident in the combinatorial mutants (white arrowheads), yet second-arch expression was not restored (double arrowheads). dlx2a (green) labels arch NCCs, and dashed lines in (J) indicate arch boundaries. (K) Ectopic ventral expression of jag1b (magenta) and prrx1b (green) in edn1 mutants (white arrows) is reversed in the mandibular (asterisk) but not hyoid (double arrowheads) domains of nr2f1b; nr2f2; nr2f5; edn1 mutants. White outlines depict the first endodermal pouch. Scale bars in (E) 50 μm, (H–K) 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
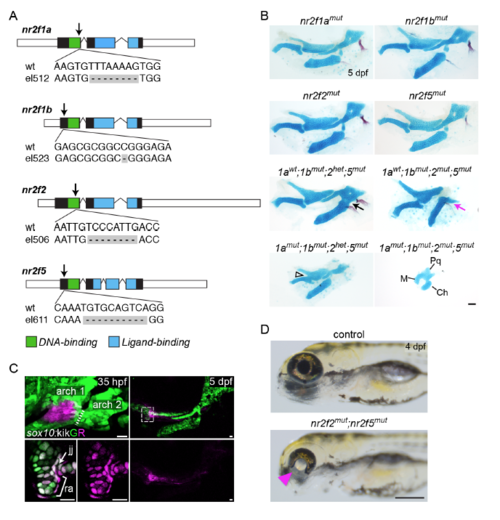
Craniofacial and ocular phenotypes in Nr2f mutants. (A) Nr2f deletion alleles generated in this study. Care was taken to introduce early frame-shift mutations likely abrogating all protein function. The mutation sites in each gene are indicated with arrows. (B) Alcian blue and Alizarin red staining of nr2f1a, nr2f1b, nr2f2, and nr2f5 single mutants at 5 dpf revealed no gross skeletal abnormalities. Combinatorial loss of nr2f1b, nr2f2, and nr2f5 alleles led to fusion of the hyoid joint (black arrow) and reduction of the dorsal hyomandibula cartilage and opercle bone (magenta arrow). Loss of nr2f1a in nr2f5 mutants deficient for nr2f1b and nr2f2 in some cases led to syngnathia (open arrowhead). The full facial skeleton is reduced in quadruple mutants. The probable identities of residual fused cartilages in the quadruple mutant are indicated. M, Meckel’s cartilage; Pq, palatoquadrate; Ch, ceratohyal. (C) First arch origins of the retroarticular process of Meckel’s cartilage in zebrafish. (Left panel) sox10:kikGR+ cells in the center of the first arch were photoconverted from green to red (magenta) fluorescence by selective exposure to UV light at 35 hpf. The right panel shows the same fish at 5 dpf, with residual photoconverted protein (magenta, also see bottom right panel) in cells within the retroarticular process (ra) and jaw joint (jj) region. The boxed area is magnified in the bottom left images. Images are maximum intensity projections except the high magnifications, which are single optical sections. (D) Coloboma in nr2f2; nr2f5 mutants (magenta triangle) was fully penetrant and allowed for straightforward identification of double mutants as early as 30 hpf. Scale bars: B = 50 μm, C = 20 μm, D = 250 μm |
|
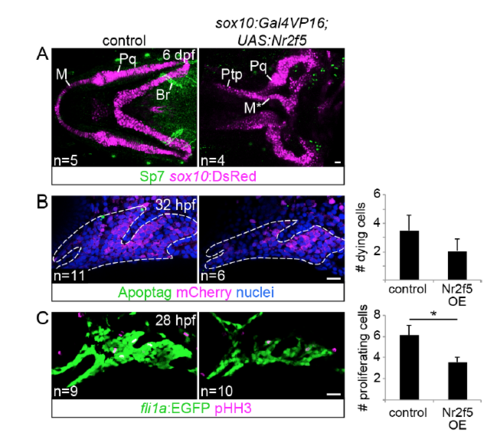
Analysis of osteoblast differentiation, cell death, and proliferation in nr2f5 misexpression embryos. (A) The sox10:DsRed transgene (magenta) and Sp7 antibody (green) label chondrocytes and osteoblasts, respectively. Note the strong Sp7 staining in the second arch-derived branchiostegal ray bone (Br). No ectopic Sp7 staining was detected in the vicinity of the transformed Meckel’s (M) cartilage. Pq, palatoquadrate; Ptp, pterygoid process (out of view in control image). (B) Similar numbers of apoptotic cells (green) were seen in the first two arches of control and sox10:Gal4VP16; UAS:nr2f5 transgenic embryos (nr2f5 OE) at 32 hpf (Mann Whitney U, p > 0.05). Arch NCCs express sox10:DsRed (mCherry antibody staining, magenta), and nuclei were labeled with Hoechst (blue). Dashed lines indicate arch boundaries. (C) nr2f5-misexpressing embryos showed a marginally significant decrease in the number of dividing cells (pHH3+, magenta) in the first two arches relative to controls (Mann Whitney U, p < 0.05). Arch NCCs express fli1a:EGFP (GFP antibody staining, green). The number of embryos evaluated for each genotype is indicated at the bottom left. Scale bars = 20 μm. |
|
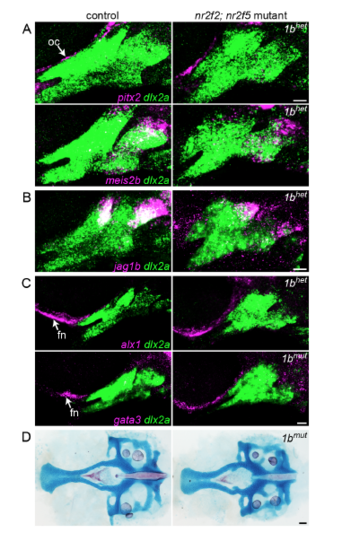
Additional gene expression patterns in nr2f2; nr2f5 mutants. (A) pitx2 and meis2b (magenta) were classified as Nr2f-activated genes by RNAseq but were not consistently altered in nr2f2; nr2f5 mutants. pitx2 was minimally expressed in dlx2a+ arch NCCs (green) but detected in the adjacent ocular mesenchyme (oc). meis2b was mainly confined to the hyoid arch. (B) jag1b expression (magenta) in non-maxillary dorsal cells was largely unaffected in Nr2f mutants. (C) Frontonasal (fn) NCC expression of alx1 and gata3 (magenta) was preserved in nr2f2; nr2f5 mutants. (D) The neurocranium, including the NCC-derived anterior portion, was slightly smaller but otherwise typically formed in nr2f2; nr2f5 mutants. Scale bars: A-C = 20 μm, D = 50 μm. |
|
Nr2f expression in fish with loss or gain of Edn1. Two-color images corresponding to the Nr2f in situ hybridizations from Fig. 5. The dashed lines in Fig. 5 were drawn based on labeling of all arch NCCs (green) by dlx2a. Nr2f expression (magenta) is elevated to varying degrees in the mandibular domain of edn1 mutants, (white arrows), whereas Edn1 misexpression induced by heat-shocking hsp70l:Gal4; UAS:Edn1 embryos from 20-24 hpf abolished most arch Nr2f expression. Scale bar = 20 μm. |
|
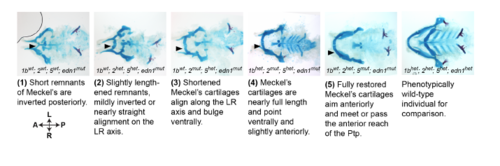
Scoring system for Meckel’s rescue. The degree of lower jaw rescue in nr2f; edn1 mutants was scored as 1 (typical edn1 mutant) through 5 (typical wild-type) as described. Representative Alcian blue and Alizarin red-stained skeletons and their associated genotypes are shown. |
Reprinted from Developmental Cell, 44(3), Barske, L., Rataud, P., Behizad, K., Del Rio, L., Cox, S.G., Crump, J.G., Essential Role of Nr2f Nuclear Receptors in Patterning the Vertebrate Upper Jaw, 337-347.e5, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell