- Title
-
Otx but not mitf transcription factors are required for zebrafish retinal pigment epithelium development
- Authors
- Lane, B.M., and Lister, J.A.
- Source
- Full text @ PLoS One
|
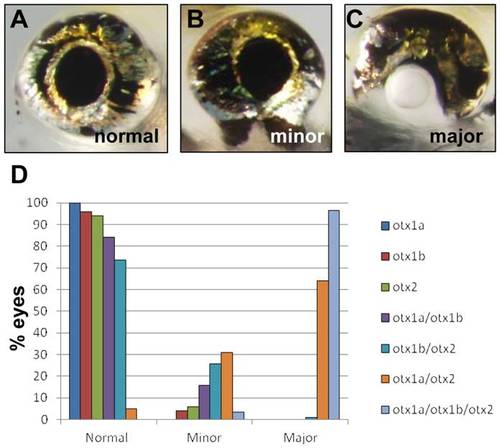
Otx morpholino knockdown results in a variable degree of RPE deficits in zebrafish. (A–B) Control morpholino (A) and otx1a/otx2 morpholino (B) injected embryos photographed at 35 hpf. RPE pigmentation is reduced in otx1a/otx2 morphants (black arrows) while neural crest derived melanocytes are unaffected (white arrows). (C–D) Ventral view of 5 dpf control morpholino (C) and otx1a/otx2 morpholino (D) injected larvae. The phenotype of otx1a/otx2 morphants can vary in severity between eyes of the same embryo, with one eye displaying a minor coloboma phenotype (black arrow) and the other eye displaying a major coloboma phenotype (white arrow). |
|
Otx morphant phenotypes. (A–C) Eye phenotypes, assessed at 5 dpf, were scored as normal when the RPE displayed no visible defects (A), minor when coloboma was present but anterior and posterior ventral RPE were still contiguous (B), and major when the anterior and posterior ventral RPE were completely separated (C). (D) Histogram showing the eye defects induced by otx gene knockdown when injected at a morpholino concentration of 1.25 ng/embryo. Knockdown of individual otx genes caused only minor defects, while knockdown of otx1a and otx2 together, or all three otx genes together, produced a majority of embryos with major eye defects. otx1a, N = 108; otx1b, N = 230; otx2, N = 122; otx1a/otx1b, N = 222; otx1b/otx2, N = 256; otx1a/otx2, N = 214; otx1a/otx1b/otx2, N = 178. PHENOTYPE:
|
|
The loss of ventral RPE in otx morphants has little effect on retinal lamination. (A) Methylene blue stained sagittal section of an otx1a/otx2 morphant classified with a minor eye phenotype. The laminar structure of the retina is unaffected despite extensive absence of RPE. (B) Zpr1 staining reveals that red-green double cone photoreceptors differentiate normally despite the loss of RPE (white arrow). Scale bar is 25 micrometers. PHENOTYPE:
|
|
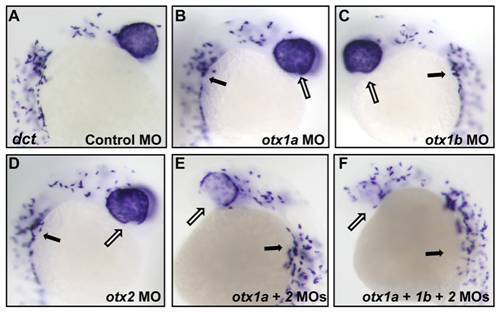
Otx knockdown causes a decrease in dct expression in the developing RPE. dopachrome tautomerase (dct) mRNA expression was analyzed through in situ hybridization at 24 hpf in otx morphants. dct expression was reduced only in the ventral most region of the optic cup in single morphants (B–D, white arrows) compared to controls (A). dct expression was significantly decreased in the ventral RPE of otx1a/otx2 morphants and to an even greater degree in otx1a/otx1b/otx2 morphants (E–F, white arrows). dct expression in neural crest melanocytes was unaffected (black arrows). Approximately 50 larvae were examined for each condition. EXPRESSION / LABELING:
|
|
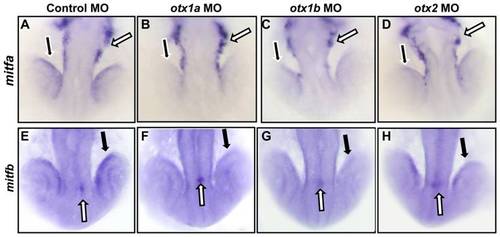
mitfa and mitfb expression is decreased in otx morphants. Expression of mitfa (A–D) and mitfb (E–H) was examined through in situ hybridization at the 21 somite stage in embryos injected with a control morpholino (A,E), or morpholinos against otx1a (B,F), otx1b (C,G), or otx2 (D,H). In situ hybridization analysis revealed a decrease in mitfa expression in the developing RPE cells (black arrows) but not in the neural crest cells (white arrows) of otx single morphants (D) and when compared to controls (A). mitfb expression was also reduced specifically in the RPE (black arrows) of otx morphants (F–H) compared to the controls, leaving epiphysis expression unaffected (white arrows). All morphants were processed simultaneously with approximately 50 larvae for each condition and the experiment was repeated twice. EXPRESSION / LABELING:
|
|
Elimination of mitfa and mitfb activity has no detectable effect on RPE development in zebrafish embryos. |
|
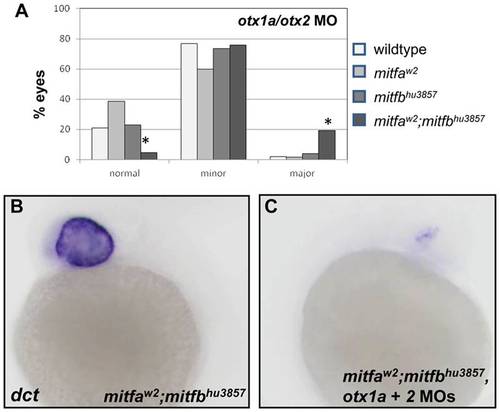
The combined knockdown of Otx and Mitf transcription factors creates a more severe phenotype. (A) The effect of otx1a/otx2 knockdown was compared between wildtype, mitfaw2 and mitfbhu3857 single mutants, and mitfbhu3857;mitfaw2 double mutants. Morpholinos were injected at a total concentration of 2 ng/embryo using the same needle for all genotypes. Results from one representative experiment are shown. The percentage of severe RPE defects was significantly increased (P<0.0001) in the mitfbhu3857;mitfaw2 double mutant background compared to wildtype and single mitfa and mitfb mutants. Wildtype, N = 252; mitfa, N = 196; mitfb, N = 158; mitfa;mitfb, N = 170. (B,C) At 24 hpf dct expression is nearly eliminated in mitfbhu3857;mitfaw2 embryos injected with the otx1a/otx2 morpholino combination (C) compared to sibling embryos injected with control morpholino (B). EXPRESSION / LABELING:
|
|
otx1a and otx2 expression is not altered in mitfa and mitfb mutants. (A–D) No difference in otx1a expression was revealed through in situ hybridization in wildtype (A), mitfaw2 (B), mitfbhu3857 (C), and mitfbhu3857;mitfaw2 (D) embryos at 21 hpf. (E–H) Likewise, otx2 expression was unchanged in wildtype (E), mitfaw2 (F), mitfbhu3857 (G), and mitfbhu3857;mitfaw2 (H) embryos at the same stage. All mutants were processed simultaneously with approximately 50 larvae examined for each condition and the entire experiment was repeated. |
|
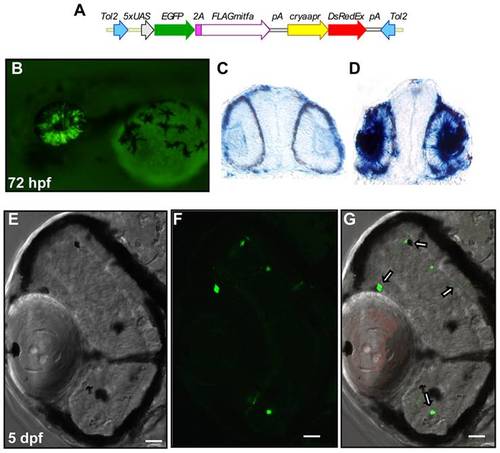
The misexpression of mitfa in the developing retina is capable of inducing foci of pigmentation. (A) Schematic of the Tol2 transposon construct expressing EGFP and FLAG-tagged Mitfa, linked by a viral 2A peptide, from a 5×UAS promoter, and containing an alpha crystallin promoter:DsRed-Express transgene marker. (B) 72 hpf larva doubly transgenic for s1003t (retinal Gal4-VP16) and UAS:EGFP-2A-FLAG-mitfa, expressing GFP in the developing retina. (C,D) 20 micron cryosections of 28 hpf larvae examined for mitfa expression through in situ hybridization revealed mitfa expression in the developing retina and lens of a GFP expressing embryo (D) but not in an embryo lacking GFP expression (C). (E–G) cryosections of a 5 dpf s1003t; UAS:EGFP-2A-FLAG-mitfa transgenic larva, displaying foci of pigmentation (E) and GFP expression (F) in the retina. (G) Overlay demonstrating extensive overlap of GFP expression and ectopic pigmentation (white arrows). Scale bar indicates 20 micrometers in (E–G). Expression of DsRed-Express is also visible in the lens in (G). |
|
Otx knockdown leads to a decrease of key RPE related genes. (A–F) Expression of silvb was examined in otx morphants at 24 hpf and representative images are shown. At 24 hpf, a small percentage of the single otx1a (B), otx1b (C) and otx2 (D) morphants displayed a slight loss of silvb expression in the ventral eye when compared to controls (A). Combined otx1a/otx2 morphants showed a more severe reduction (E) and expression was almost completely eliminated in otx1a/otx1b/otx2 morphants (F). |