PUBLICATION
Hypoxia deactivates epigenetic feedbacks via enzyme-derived clicking proteolysis-targeting chimeras
- Authors
- Do, T.C., Lau, J.W., Sun, C., Liu, S., Kha, K.T., Lim, S.T., Oon, Y.Y., Kwan, Y.P., Ma, J.J., Mu, Y., Liu, X., Carney, T.J., Wang, X., Xing, B.
- ID
- ZDB-PUB-221216-13
- Date
- 2022
- Source
- Science advances 8: eabq2216eabq2216 (Journal)
- Registered Authors
- Keywords
- none
- MeSH Terms
- none
- PubMed
- 36516252 Full text @ Sci Adv
Abstract
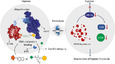
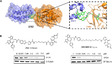
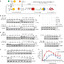
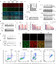
Epigenetic mediation through bromodomain and extraterminal (BET) proteins have progressively translated protein imbalance into effective cancer treatment. Perturbation of druggable BET proteins through proteolysis-targeting chimeras (PROTACs) has recently contributed to the discovery of effective therapeutics. Unfortunately, precise and microenvironment-activatable BET protein degradation content with promising tumor selectivity and pharmacological suitability remains elusive. Here, we present an enzyme-derived clicking PROTACs (ENCTACs) capable of orthogonally cross-linking two disparate small-molecule warhead ligands that recognize BET bromodomain-containing protein 4 (BRD4) protein and E3 ligase within tumors only upon hypoxia-induced activation of nitroreductase enzyme. This localized formation of heterobifunctional degraders promotes specific down-regulation of BRD4, which subsequently alters expression of epigenetic targets and, therefore, allows precise modulation of hypoxic signaling in live cells, zebrafish, and living mice with solid tumors. Our activation-feedback system demonstrates compelling superiorities and may enable the PROTAC technology with more flexible practicality and druggable potency for precision medicine in the near future.
Genes / Markers
Expression
Phenotype
Mutations / Transgenics
Human Disease / Model
Sequence Targeting Reagents
Fish
Orthology
Engineered Foreign Genes
Mapping