- Title
-
FARS2 Deficiency Causes Cardiomyopathy by Disrupting Mitochondrial Homeostasis and the Mitochondrial Quality Control System
- Authors
- Li, B., Liu, F., Chen, X., Chen, T., Zhang, J., Liu, Y., Yao, Y., Hu, W., Zhang, M., Wang, B., Liu, L., Chen, K., Wu, Y.
- Source
- Full text @ Circulation
|
|
|
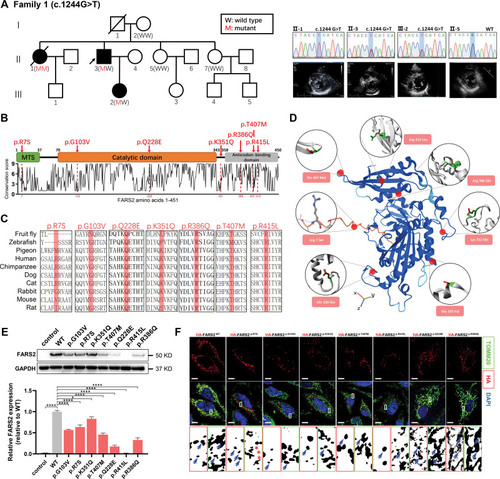
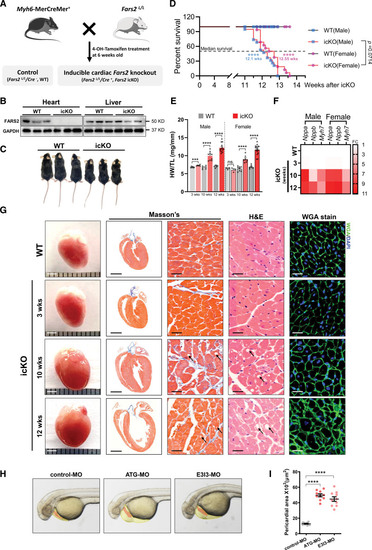
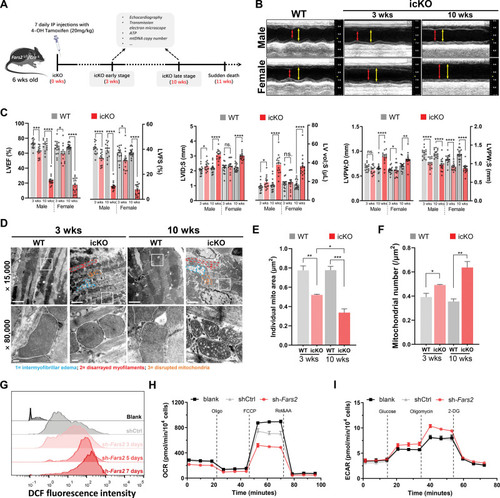
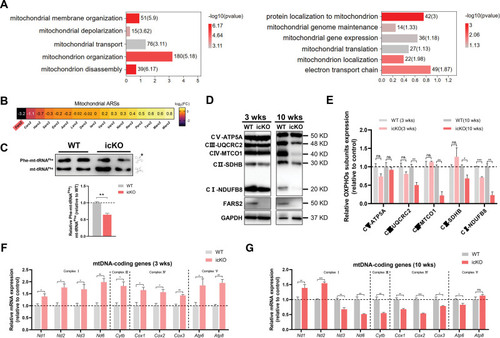
PHENOTYPE:
|
|
|
|
|
|
|
|
|
|
|
|
|

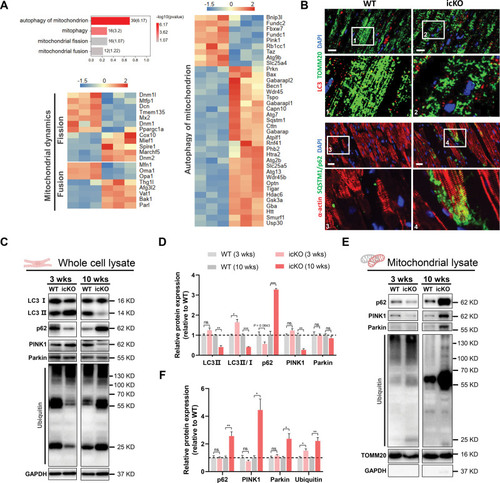
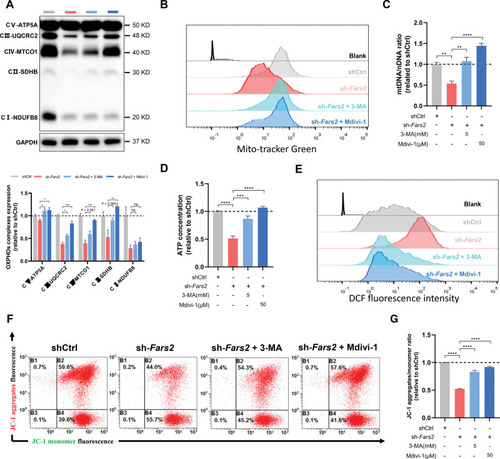
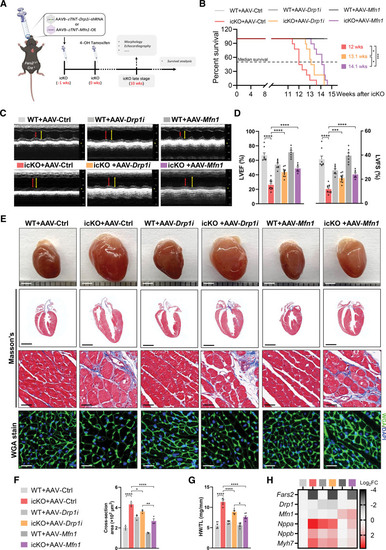
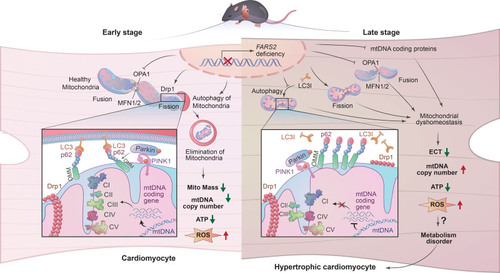
Unillustrated author statements PHENOTYPE:
|