- Title
-
Canonical Wnt and TGF-β/BMP signaling enhance melanocyte regeneration but suppress invasiveness, migration, and proliferation of melanoma cells
- Authors
- Katkat, E., Demirci, Y., Heger, G., Karagulle, D., Papatheodorou, I., Brazma, A., Ozhan, G.
- Source
- Full text @ Front Cell Dev Biol
|
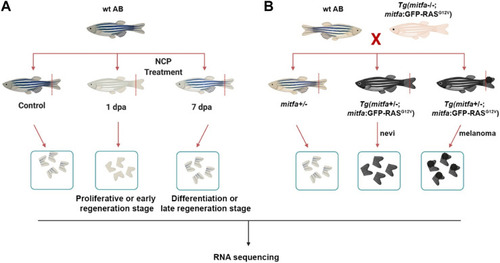
Generation of the zebrafish models of melanocyte regeneration and melanoma |
|
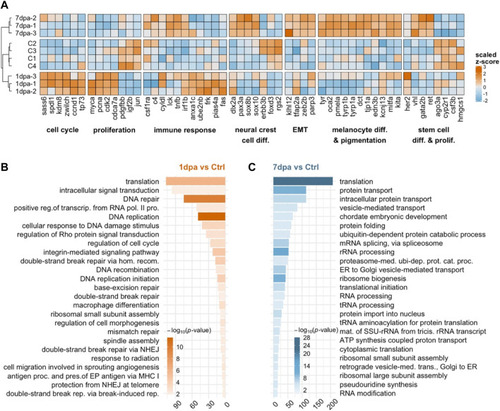
Validation of the melanocyte regeneration model and quantitative analysis of the DEGs at the proliferation and differentiation stages |
|
Early/proliferative and late/differentiation stages of melanocyte regeneration have distinct transcriptional profiles |
|
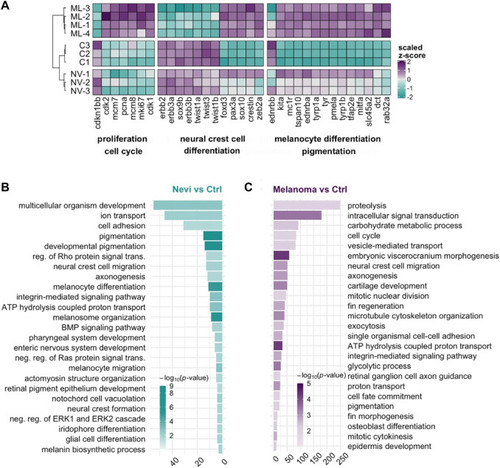
Validation of the melanoma model and quantitative analysis of the DEGs at the nevi and melanoma stages |
|
Transcriptional profile of melanoma differs from that of nevi by the proliferative burst and oppositely regulated NCC signature |
|
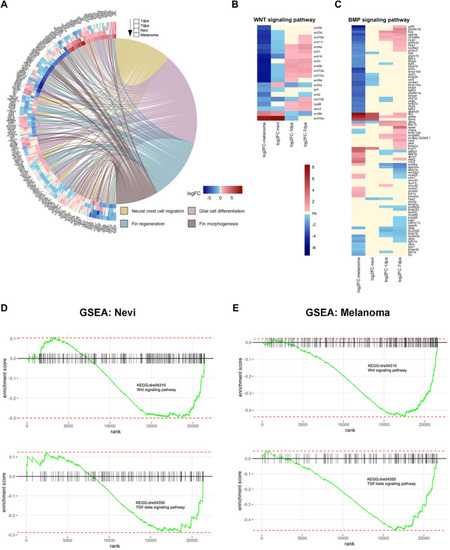
Cellular processes, including Wnt and TGF-β/BMP signaling pathways, are differentially regulated between melanocyte regeneration and melanoma |
|
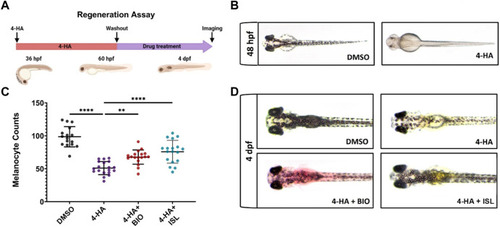
Activation of canonical Wnt or TGF-β/BMP signaling pathways enhances larval melanocyte regeneration |
|
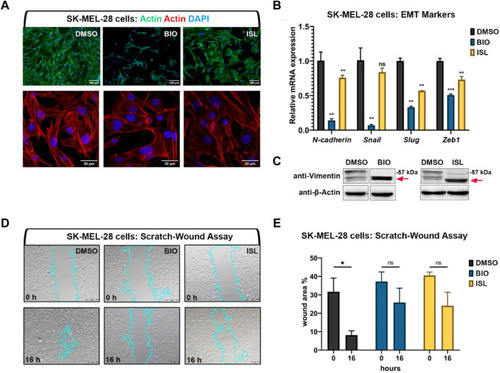
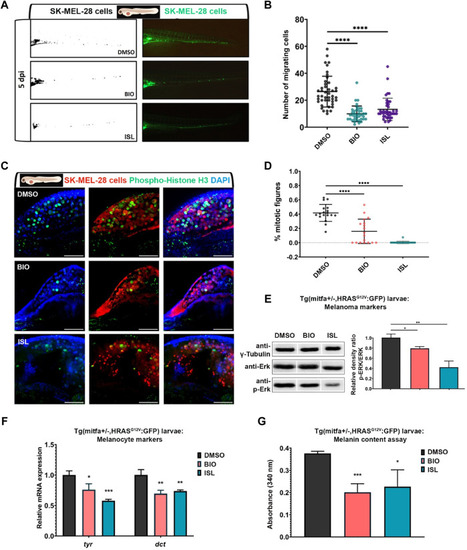
Activation of canonical Wnt or TGF-β/BMP signaling pathways suppresses invasiveness of human melanoma cells |
|
Activation of canonical Wnt or TGF-β/BMP signaling pathways suppresses migration and proliferation of human melanoma cells |