- Title
-
Dynamic spatiotemporal coordination of neural stem cell fate decisions occurs through local feedback in the adult vertebrate brain
- Authors
- Dray, N., Mancini, L., Binshtok, U., Cheysson, F., Supatto, W., Mahou, P., Bedu, S., Ortica, S., Than-Trong, E., Krecsmarik, M., Herbert, S., Masson, J.B., Tinevez, J.Y., Lang, G., Beaurepaire, E., Sprinzak, D., Bally-Cuif, L.
- Source
- Full text @ Cell Stem Cell
|
Inhibitory interactions between progenitor cells bias the distribution of NSC activation events (A–A’’') Confocal dorsal views of a whole-mount adult telencephalon showing the germinal layer of the pallium in a 3mpf (A and A’) Dorsal (apical) view and high magnification of the NSC layer, showing merged and single channels (color-coded). Arrowheads: purple to aNSCs, orange to aNPs. aNSCs are GFP+,Gfap+, aNPs are negative for both markers. (A’’ and A’’’) Examples of aNSCs with a process (red arrows) visible using the GFP or the Zrf1 markers, respectively (left and right panels: different backgrounds to ease observation). (B–B’’) Similar sample stained for GFP (green, NSCs) and PCNA (magenta, proliferating cells). (B and B’) Merged and single-channel views (color-coded). (B’’) Close-up in Dm showing progenitor cell states (merged image, individual channels and segmentation): quiescent NSCs (qNSCs; GFP+ only; green), activated NSCs (aNSCs; GFP+,PCNA+; magenta), and proliferating neural progenitors (aNPs; PCNA+ only; orange) (see also (A, B, and B') Stitches of 4 tiles with 10% overlap. (C) Main pallial NSC lineage (arrows: lineage transitions). (D) Proportions of qNSCs, aNSCs, and aNPs relative to each other in Dm (see also (E) Besag’s (F) Range and strength of this interaction determined with functions Scale: (A), (B), and (B’) 100 μm; (A’) 10 μm; (A’’), (A’’’) 7 μm; (B’’) 30 μm. |
|
The lower incidence of activated NSCs close to aNPs is Notch signaling dependent (A) Confocal whole-mount view of the pallial germinal layer in a 3mpf (B–B’’’) Close-ups in Dm, all channels. Arrowheads: magenta, aNSC; orange, aNP; expressing (C–C’’) Percentages of aNSCs (all, pre-, or post-division) among all NSCs (C and C”) or aNSCs (C’) upon 24-h LY treatment (DMSO: control). ns, non-significant; unpaired t test (see also (D–D”) Number (D and D’) and proportion (D’’) of aNSCs, in total (D) or touching aNPs upon 24-h LY treatment (DMSO: control) (D’’, p = 0.002, t test) (see also (E) Spatial distribution (Besag’s Scale: (A) 40 μm, (B)–(B’’’) 20 μm. |
|
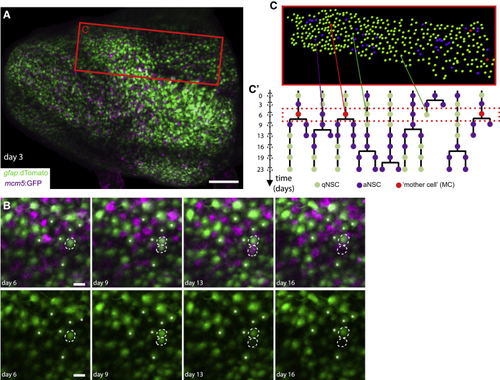
Intravital imaging resolves adult NSC lineage trees in time and space (A) Whole pallial hemisphere imaged intravitally in a 3mpf (B) Close-ups from the same video showing an asymmetric NSC division (dotted circles) between days 3 and 9: 1 daughter differentiates over the next 7 days (bottom dotted circle, loss of the (C and C’) Segmentation and NSC tracking over 23 days in Dm in Mimi. (C) Segmentation of ~390 cells per time point (area boxed in A, at day 6). (C’) Example of dividing tracks, with cell states (color-coded) and the spatial position of each tree (see also |
|
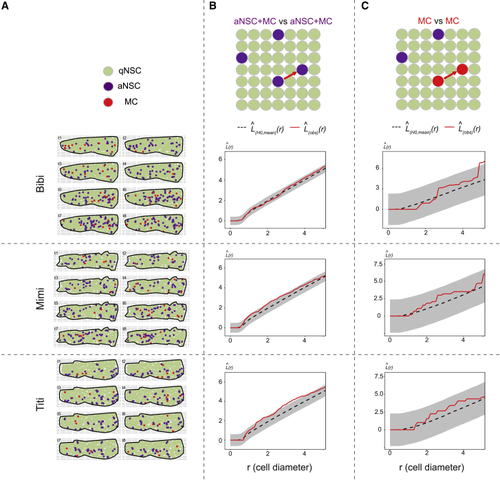
Static spatial analysis of NSC activation from dynamic datasets (A) Dm surfaces segmented for the 3 fish analyzed (Bibi, Mimi, Titi) at all time points, cell states color-coded (see also (B and C) |
|
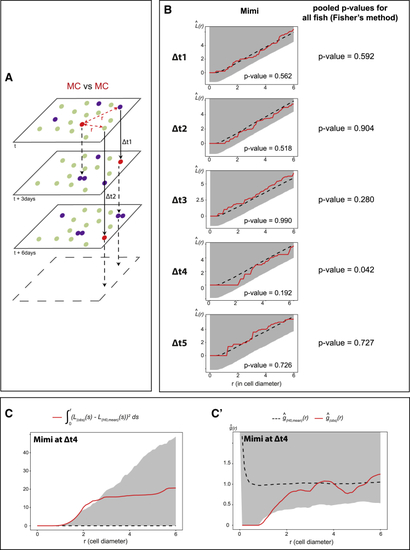
NSC division events are spatiotemporally coordinated (A) MC positions at time point t are compared, using point pattern statistics, with MC positions at other time points after fixed intervals (t+Δt), for each possible Δt (3, 6, 9, and 15 days). (B) Left: Besag’s (C and C’) Illustrations for Mimi. (C) Integrated square of the negative deviation between |
|
An NSC lattice model captures NSC population dynamics (A) Model lineage flowchart. γ: transition rates; Pqq, Pqp, and Ppp: probabilities for (as)symmetric aNSC divisions; dashed line: lateral inhibition (LI) of aNPs on NSC activation (see also (B and B’) Snapshot of the lattice (B) and (B’) examples of activation (purple arrowhead), division (black star), and differentiation (orange arrowhead) ( (C and D) Average cell proportions (C) and (D) percentages of cell fates following cell division in sets of 18 simulations with and without LI versus (E–E’’’) Dynamics and spatial correlations from the NSC lattice model with (top) and without (bottom) aNP-driven LI. (E) Stable cell numbers and proportions over time, matching experimental data. Bold line: mean; shaded area: range; n = 18 simulations.. (E’) Snapshots of the lattice at 1 time step from a simulation of 500 time steps. Red arrowheads: aNSCs neighboring aNPs. (E’’ and E’’’) Besag’s |
|
The NSC lattice model reproduces the spatiotemporal correlations of NSC fate decisions and shows their impact on long-term neuronal distribution (A) Top: MC positions at any time t in the lattice model are compared using point pattern statistics with MC positions at t+Δt. Bottom left: Besag’s (B) Compared (C) Compared spatial interaction between output neurons in a model with (pink) and without (blue) LI, using a 3D Ripley’s (D) Similar comparison (to C) in 2D, using the (B)–(D) show centered summary functions, |