- Title
-
A single-cell resolution developmental atlas of hematopoietic stem and progenitor cell expansion in zebrafish
- Authors
- Xia, J., Kang, Z., Xue, Y., Ding, Y., Gao, S., Zhang, Y., Lv, P., Wang, X., Ma, D., Wang, L., Han, J.J., Liu, F.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
Single-cell transcriptome map of zebrafish CHT. (A) A schematic paradigm of tissue processing and fluorescence-activated cell sorting for scRNA-seq profiling of CHT. ECs, HCs, and NCs were sorted from Tg (kdrl:mCherry/CD41:GFP) embryos across three developmental stages (55 hpf, 3.5 dpf, and 4.5 dpf). (B) UMAP plot showing 23 CHT cell clusters across three developmental stages (28,777 cells). Cells are colored by their cell-type annotation and numbered according to the legend beside. (C) Heatmap showing blocks of DEGs (top 10 genes) in HSPCs, neural cells, erythrocytes, myeloid cells, ECs, epithelial cells, epidermis, fibroblasts, and muscle cells in the CHT. (D) UMAP visualization of the expression of curated feature genes for cell-type identification. |
|
Cell composition and gene-expression dynamics during CHT development. (A) Fraction of cell clusters per developmental stage, displaying a dynamic change in cell-type complexity throughout our sampling. (B) Dot plot showing the differentially expressed TFs within the HSPCs at each development stage. (C) Dot plots showing the differentially enriched GO terms within HSPCs, ECs, neural cells, and fibroblasts at each developmental stage. PHENOTYPE:
|
|
HSPC heterogeneity in lineage priming and metabolic gene signatures. (A) Dot plot of 14 selected DEGs for HSPC 1 through 4 and blood lineages (erythrocytes 1/2, macrophage, and monocyte 2). The size of the dot corresponds to the percentage of cells expressing the gene in each cluster. The color represents the average expression level. (B) The major GO terms enriched in HSPC 1 through 4. (C) PAGA visualization layout of HCs derived from HSPCs (15,084 cells). PAGA showing the putative developmental process from the root state (HSPC 1) to the branches (erythrocyte, myelocyte, and HSPC 4) passing through HSPC 2 and 3. (D) The fraction of four HSPCs at each development stage. (E) Heatmap showing the dynamic gene expressions based on PAGA-inferred trajectories from HSPC 1 to erythrocytes, HSPC 4, and macrophages, respectively. (F) The major metabolic pathway–activity score in HSPC 1 through 4. (G) Violin plots showing the expression of representative metabolic marker genes for cell cycle, ferroptosis, apoptosis, endocytosis, lysosome, and phagosome in HSPC 1, HSPC 2, HSPC 3, and HSPC 4. |
|
Cell–cell interaction network between HSPCs and nonhematopoietic niche components. (A) The interaction network of TFs of each component. All differentially expressed TFs in each cell type are used to construct network; nodes (TFs) with more than one edge are shown. (B) Dot plot showing the frequency of ligand–receptor genes involved in Kyoto Encyclopedia of Genes and Genomes pathways. (C) Dot plot showing the ligand (red)–receptor (black) pairs between HSPCs and ECs, EC-like cells, fibroblasts, neural cells, epidermal cells, and epithelial cells. (D) Violin plot showing the normalized expression of the ligand and its receptor genes in HSPCs and ECs, fibroblasts, neural cells, spinal cord cells, epidermis, and epithelial cells for each indicated pairing. (E) WISH indicating the expression patterns of ccr9a, notch2, and ptprr in HSPCs, ccl25b and dll4 in ECs, and fgfr2 in fibroblasts at 3.5 dpf. (Scale bar, 100 μm.) EXPRESSION / LABELING:
|
|
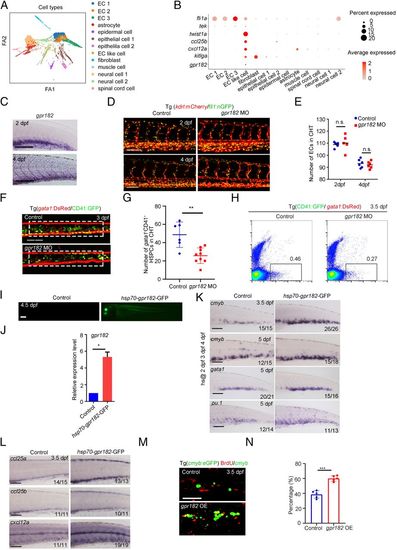
Identification of crucial niche regulator for HSPC expansion. (A) ForceAtlas2 plot of nonhematopoietic niche cells (10,200 cells). Cells are colored by their cell-type annotation. (B) Dot plot showing the expression of selected known EC, muscle cell, and epithelial cell genes (fli1a, tek, twist1a, ccl25b, cxcl12a, kitlga, and gpr182) for niche cell clusters. (C) WISH showing the expression of gpr182 in CHT at 2 dpf and 4 dpf. (D) Confocal imaging of the Tg (kdrl:mCherry/fli1:nGFP) showing the structure of ECs in the CHT region at 2 dpf and 4 dpf in control and gpr182 morphants. (Scale bar, 50 mm.) (E) The statistical data of the CHT-EC number in control and morphants at 2 dpf and 4 dpf. The dashed boxes indicate the region of EC counting. (F) Confocal imaging of the Tg (CD41:GFP/gata1:DsRed) showing the number of CD41 +gata1− HSPCs in the CHT region at 3 dpf in control and gpr182 morphants. (Scale bar, 50 mm.) (G) The statistical data of the CHT CD41 +gata1− HSPC number in F. The dashed boxes indicate the region of CD41+gata1− HSPC counting. (H) Fluorescence-activated cell sorting analysis showing the number of CD41+ gata1− HSPCs in control and gpr182 morphants at 3.5 dpf. (I) The imaging of GFP fluorescence in Tg (hsp70-gpr182-GFP) embryos at 4.5 dpf. Heat shock was performed at 2 dpf, 3 dpf, and 4 dpf. (J) qPCR result of gpr182 expression in control and hsp70-gpr182-GFP-positive embryos. (K) WISH showing the expression of cmyb, gata1, and pu.1 in the CHT of the control and hsp70-gpr182-GFP-injected embryos at 3.5 dpf and 5 dpf. (L) WISH showing the expression of ccl25a, ccl25b, and cxcl12a in control and gpr182-overexpressed embryos at 3.5 dpf. (M) The double staining imaging of anti-BrdU and anti-GFP antibodies in control and gpr182-overexpressed embryos at 3.5 dpf. OE, overexpression. (N) The statistical data of the percentage of cmyb:GFP+BrdU+/cmyb:GFP+ cells in M. (Scale bar (C, I, L, and M), 100 μm.) The results are represented as means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Student’s t test. |
|
EC-specific Gpr182 is required for HSPC expansion in mouse FL. (A) The expression pattern of Gpr182 in mouse FL scRNA-seq data. (B) qPCR analysis of Gpr182 in ECs, LSK cells, and NC cells. Lyve1 and Hlf as positive markers for ECs and HSPCs, respectively. (C) The flowchart of ex-vivo siGpr182 knockdown and HSPC culture. (D) CFU-cell assays to detect the colony-forming ability of HSPCs following siRNA treatment. |
|
Cross-species analysis between human FL and zebrafish CHT reveals conserved and divergent gene-expression profiles. (A, Left) Unsupervised graph-based clustering of single cells from human FL (n = 8,432) and zebrafish CHT (n = 10,010) based on homologous gene-expression profiles; color labels for different species. (A, Right) Color labels for identified cell clusters. (B) Similarity matrix showing the Pearson correlations between each pair of corresponding cell clusters from human FL and zebrafish CHT based on homologous gene-expression profiles. (C) Dot plot showing the conserved signature genes of each cluster in human and zebrafish. (D) Identification of GPR182, DLL4, and NOTCH2 expression in human FL cell clusters. (E) The major GO terms of divergent HSPC genes in human FL and zebrafish CHT. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |