- Title
-
Lef1 is required for progenitor cell identity in the zebrafish lateral line primordium
- Authors
- McGraw, H.F., Drerup, C.M., Culbertson, M.D., Linbo, T., Raible, D.W., and Nechiporuk, A.V.
- Source
- Full text @ Development
|
Abnormal primordium patterning leads to loss of terminal neuromasts in the lef1nl2 mutant. (A,B) eya1 expression in the pLL of wild-type sibling and lef1nl2 mutant embryos at 2 dpf. The lef1nl2 mutant lacks a tc. (A′,B′) Distal limit of eya1 expression in wild-type and lef1nl2 mutant (arrow) embryos. (C) Average number of NMs (excluding tc) in lef1nl2 mutant and wild-type sibs (mean±s.d.) are not significantly different at 2 dpf (n=28 wild type, 11 lef1nl2, P=0.49, Student′s t-test). (D) Axial positions of L1-L5 in lef1nl2 and wild-type siblings at 2 dpf (mean±s.d.). The axial level of stalled primordium (prim) in lef1nl2 mutants is indicated by the pink bar. Position of L1-L5 is significantly shifted anteriorly in lef1nl2 mutants (n=18, P<0.001, two-way ANOVA with replication). (E-F′′) Stills from time-lapse movies of primordium migration in wild-type sibling and lef1nl2 mutant embryos that express the Tg(–8.0cldnb:lynGFP) transgene. Wild-type and lef1nl2 embryos were imaged beginning at 34 hpf for 780 minutes and 840 minutes, respectively (see Movies 1 and 2 in the supplementary material). (E-E′′) Over the course of 780 minutes, the primordium in the wild-type embryo migrated out of frame, deposited three NMs (red, blue and yellow asterisks) and generated two new rosettes (green and pink asterisks). (F-F′′′) Over the course of 840 minutes, the primordium in the lef1nl2 mutant has slowed and became elongated (pink asterisk), having deposited four NMs (red, blue, yellow and green asterisks). Scale bars: 20 μm. |
|
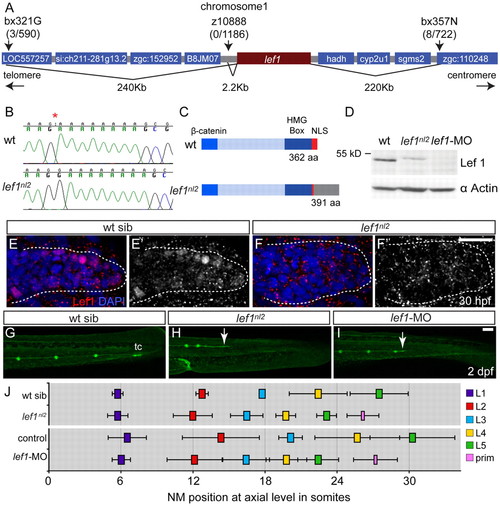
The lef1nl2 mutant contains a lesion in the lef1 gene. (A) The lef1nl2 mutation was mapped to a 460 Kb region on chromosome 1. Numbers of recombinants are indicated under each marker. (B) The lef1nl2 mutation is a single guanine insertion at position 1120 (red asterisk). (C) The resulting frame shift is predicted to disrupt the nuclear localization signal (NLS) and extend the protein by 29 amino acids (aa; also shown a β-catenin binding domain and HMG Box). (D) Western blot of wild-type, lef1nl2 mutant and lef1 morphant whole embryo lysates probed with anti-Lef1 antibody and anti-α actin antibody. (E-F′) Immunolabeling using anti-Lef1 antibody revealed nuclear labeling (red) in wild-type sibling and cytoplasmic labeling in lef1nl2 mutant embryos. Nuclei are labeled with DAPI. (G-I) Wild-type sibling, lef1nl2 mutant and lef1-MO injected embryos expressing the Tg(–8.0cldnb:lynGFP) transgene at 2 dpf. (G) The pLL was truncated prematurely (white arrows) in the lef1nl2 mutant (H) and lef1 morphant (I). (J) The axial positions of the deposited NMs are shifted anteriorally in lef1nl2 mutants when compared with wild-type siblings and in lef1 morphants when compared with uninjected controls. (Data presented as mean±s.d.; n=6-11 P<0.001, two-way ANOVA with replication.) Scale bars: 50 μm. |
|
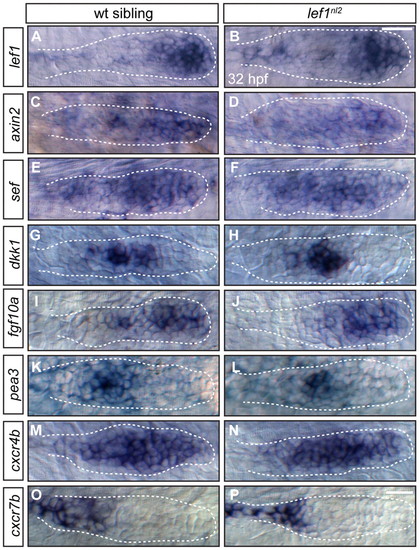
Primordium patterning is maintained in lef1nl2 mutants. (A-P) RNA in situ hybridization of factors required for primordium patterning in wild-type siblings (left panels) and lef1nl2 mutants (right panels) at 32 hpf. Expression of lef1 (A,B), axin2 (C,D), sef (E,F), dkk1 (G,H), fgf10a (I,J), pea3 (K,L), cxcr4b (M,N) and cxcr7b (O,P) are similar in both wild-type and mutant embryos. Scale bars: 20 μm. EXPRESSION / LABELING:
|
|
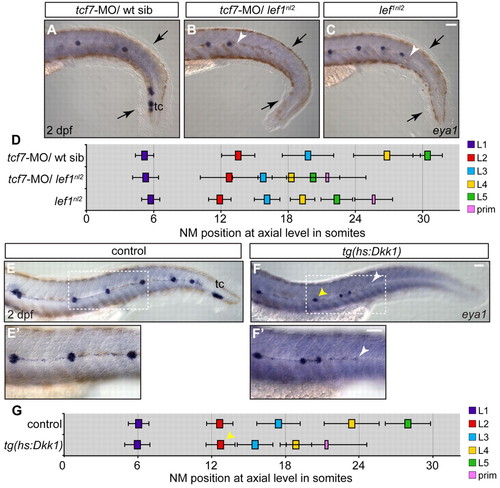
Combined loss of Lef1 and Tcf7 does not recapitulate the pLL phenotype seen with the complete loss of Wnt signaling. (A-C) Zygotes from a lef1nl2/+ incrosses were injected with tcf7-MO. NM distribution in the injected embryos was assessed by eya1 expression at 2 dpf and compared with uninjected lef1 mutant embryos. The loss of Tcf7 alone did not affect pLL patterning in lef1nl2/+ sibling (A), the pLL is truncated more severely lef1nl2/–; tcf7-MO (B; arrowhead) compared with lef1nl2 mutants alone (C; arrowhead). Note reduced fin fold (black arrows) caused by a combined loss of Lef1 and Tcf7 as reported previously (Nagayoshi et al., 2008). (D) Axial positions of the pLL NMs (L1-L5) and primordium (mean±s.d.; n=11, P<0.001, two-way ANOVA with replication). (E-F′) pLL formation in 2 dpf embryos following the induction of Dkk1 in the Tg(hsp70l:dkk1-GFP) line by a heat-shock at 28 hpf. Position of the primordium at the time of heat-shock is marked by the yellow arrowheads. Dashed rectangles indicate the regions shown at higher magnification in E′,F′. Following the heat-shock, one or two NMs are deposited and the pLL ends with a trail of eya1-positive cells (white arrowheads). (G) Axial positions of NMs deposited following heat-shock in control and Tg(hsp70l:dkk1-GFP) embryos (n=11). Scale bars: 20 μm. |
|
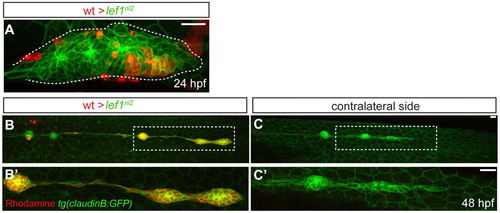
Lef1 function is required in leading edge cells of the primordium for proper pLL formation. (A-C′) Confocal projection obtained from a lef1nl2 mutant host that received wild-type donor cells (rhodamine dextran, red). (A) At 24 hpf, the primordium contains donor cells in the leading zone and caudal-most rosette. (B) The same embryo as in A, showing complete primordium migration and tc formation at 48 hpf. (B′) High magnification of region outlined in B; the primordium is entirely composed of donor cells. (C) Lateral line is truncated on the contralateral side of the same embryo. (C′) High magnification of the region outlined in C. Both wild-type donors and mutant hosts expressed Tg(–8.0cldnb:lynGFP) transgene. Scale bars: 20 μm. |
|
Wnt signaling regulates proliferation in the primordium. (A-C′′) Confocal projections of primordia following BrdU incorporation between 32.5 and 34.0 hpf in wild-type, lef1nl2 mutant and Tg(hsp70l:dkk1-GFP) embryos. All embryos express Tg(–8.0cldnb:lynGFP). BrdU incorporation in the primordia of the wild-type (A-A22), the lef1nl2 mutant (B-B22) and the Tg(hsp70l:dkk1-GFP) transgenic embryos heat-shocked at 28 hpf (C-C22). Scale bar: 20 μm. (D,E) BrdU incorporation index for wild-type sibling and lef1nl2 mutants (D) and control and Tg(hsp70l:dkk1-GFP) embryos (E). (F,G) BrdU incorporation index for leading region in wild-type siblings and lef1nl2 mutants (F) and controls and Tg(hsp70l:dkk1-GFP) embryos (G) (n=10-22 embryos; data presented as mean±s.e.m., **P<0.009, ***P<0.002, Student′s t-test). The leading region is defined as the cells posterior to the leading rosette in the primordium. EXPRESSION / LABELING:
|
|
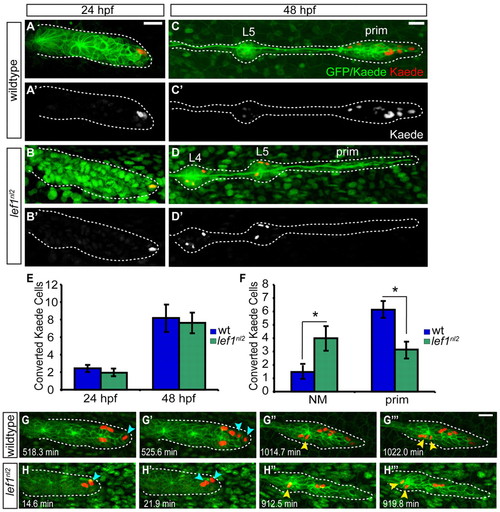
Leading region cells change their fate in the absence of Lef1 function. (A-C′) Tg(–8.0cldnb:lynGFP) zygotes were injected with a nuclear-localized Kaede mRNA and an average of two cells were photoconverted at 24 hpf. (A,B) Wild-type and lef1nl2 mutant embryos immediately following photoconversion are shown. (C-D′) The embryos in A-B′ at 48 hpf showing the location of the progeny of the photoconverted cells. Note absence of the labeled (red) cells in the leading region of lef1nl2 mutant embryos. (E) Quantification of converted cells at 24 hpf and their progeny at 48 hpf. (F) Localization of converted cells at 48 hpf in wild-type and lef1nl2 mutants. Cells in lef1nl2 mutants are significantly more likely to be localized in NMs and not in the primordia when compared with wild type (mean±s.e.m., n=27 wild-type and 16 lef1nl2 mutant embryos, *P<0.04, Student′s t-test). (G-H′′) Still images from time-lapse movies (see Movies 3 and 4 in the supplementary material) demonstrating division of Kaede-positive cells (red) in a wild-type (G-G′′) and a lef1nl2 embryos (H-H′′). Specific time points were chosen to show a subset of labeled cells just before and after cell divisions. Leading zone divisions marked by blue arrows, whereas cell divisions in rosettes are marked by yellow arrowheads. Scale bars: 20 μm. |
|
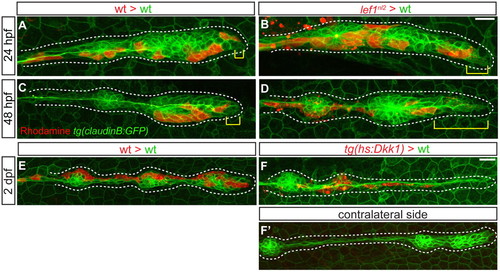
Lef1 is required to maintain progenitor cell identity. (A-D) Confocal projections of Tg(–8.0cldnb:lynGFP)-positive, mosaic embryos containing either wild-type or lef1nl2 donor cells (red) at 24 and 48 hpf. (A) Wild-type host primordium at 24 hpf with wild-type donor cells in the leading region (yellow bracket). (B) Chimeric primordium containing lef1nl2 mutant cells in the leading region at 24 hpf (yellow bracket). At 48 hpf, wild-type donor cells remain in the leading region (C; yellow bracket), whereas lef1nl2 cells have moved out of the leading region (D; yellow bracket). (E-F′) Confocal projections of chimeric primordia containing wild type Tg(hsp70l:dkk1-GFP) donor cells. Embryos were heat-shocked at 28 hpf. (E) Primordium containing wild-type cells and a characteristic rounded morphology. (F) Wild-type primordium containing Tg(hsp70l:dkk1-GFP) donor cells shows loss of primordium organization. (F′) Contralateral side of the chimera shown in F is normal. Scale bars: 20 μm. |
|
lef1nl2 mutants lack terminal NMs and have a truncated pLL nerve. (A,B) Wild-type sibling and lef1nl2 mutant larvae at 4 dpf. The pLL nerve is visualized by expression of the TgBAC(neurod:EGFP) transgene. NMs are labeled with DASPEI. (A) Wild-type embryo contains NMs L1-L5, a secondary (2°) NM and the terminal cluster (tc). (B) lef1nl2 mutant contains NMs L1-L5 and a 2° NM, but lacks a terminal cluster (tc). In addition, the lef1nl2 mutant has a truncated pLL nerve (white arrow). (C,D) Functional hair cells in the final deposited NM labeled with FM 1-43 (red) in wild-type (C) and lef1nl2 larvae (D) at 4 dpf. Nuclei are labeled with DAPI. (E) Quantification of FM 1-43-labeled hair cells showing no significant difference between wild-type and mutant larvae (F) Average number of DAPI-labeled nuclei per NM in wild-type and lef1nl2 (n=6, mean ± s.e.m.). Scale bar: 20 μm. |
|
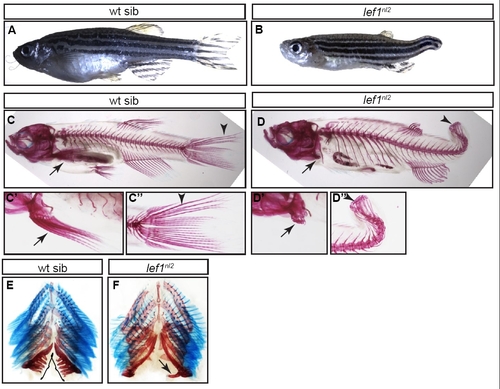
Adult lef1nl2 mutants show defects in fin and tooth formation. (A,B) Bright-field images of adult wild-type sibling (A) and lef1nl2 mutant (B) fish. Note that the lef1nl2 mutant lacks a caudal fin. (C-D′′) Alizarin Red staining in calcified bone in wild-type (C) and lef1nl2 mutant (D) fish. Lepidotrichia in both paired pectoral fins (arrows) and the caudal fin (arrowheads) are formed in the wild-type fish (C-C′′). By contrast, these structures are stunted in the lef1nl2 mutant (D-D′′). (E,F) Alizarin Red staining of the pharyngeal teeth and bone of the jaw and Alcian Blue staining of cartilage in the gill rakers. (E) Wild-type jaw with several pharyngeal teeth (black brackets). (F) By contrast, the lef1nl2 mutant jaw contains only one tooth (black arrow). Gill rakers appear normal in both wild-type and lef1nl2 mutant fish. PHENOTYPE:
|
|
Adult pLL is truncated in lef1nl2 mutants. DASPEI labeling of the pLL in adult wild-type sibling and lef1nl2 mutant fish carrying the TgBAC(neurod:EGFP)nl1 transgene. (A) The wild-type fish shows NM stitches in the lateral (white arrowheads) and ventral (red arrowhead) stripes. (B) The lef1nl2 mutant has a truncated lateral stripe and fully formed ventral stripe (blue arrow). Adults were between 4 months post-fertilization and 22-23 mm in length. PHENOTYPE:
|
|
Primordium patterning is disrupted following induction of Tg(hsp70l:dkk1-GFP). RNA in situ hybridization at 32 hpf to visualize expression of patterning markers in wild-type embryos (left panels) and Tg(hsp70l:dkk1-GFP) embryos heat-shocked at 28 hpf reveals that these factors are downregulated following Dkk1 transgene induction; lef1 (A,B), fgf10a (C,D) and pea3 (E,F). Expression of the chemokines cxcr4b (G,H) and cxcr7b (I,J) are indistinguishable between wild-type and heat-shocked Tg(hsp70l:dkk1-GFP) embryos. Scale bar: 20 μm. EXPRESSION / LABELING:
|
|
Expression of tcf genes in the primordium. At 30 hpf, tcf7 is expressed in the leading zone of the primordium (A), tcf7l1a is expressed throughout the primordium, but not in leading edge cells (B) and tcf7l1b is expressed in a deposited NM, but not in the primordium (C) in a wild-type embryo. Scale bar: 20 µm. (D) Pectoral fin development in embryos injected with the tcf7-MO in a heterozygous (E) or lef1nl2 mutant (F) background at 6 dpf. Pectoral fins are stunted or absent in the lef1nl2;tcf7-MO embryo when compared with the lef1nl wild-type sibling injected with tcf7-MO (red arrows). Scale bar: 50 μm. |
|
Loss of Lef1 function does not lead to cell death in the primordium. (A-B′′′) Confocal projections of 40 hpf wild-type (A) and lef1nl2 mutant (B) primordium that have been labeled by TUNEL for cell death (A′,B′), with Tg(-8.0cldnb:lynGFP) to visualize primordium (A′′,B′′) and by DAPI to visualize nuclei (A′′′,B′′′). There are no detectable TUNEL-positive cells in lef1nl2 mutants when compared with wild-type controls. (C-D′′′) Tunnel labeling of wild-type (C) and lef1-MO-injected (D) embryos at 35 hpf. Little TUNEL labeling was detected in lef1-MO embryos. Scale bar: 20 μm. EXPRESSION / LABELING:
|
|
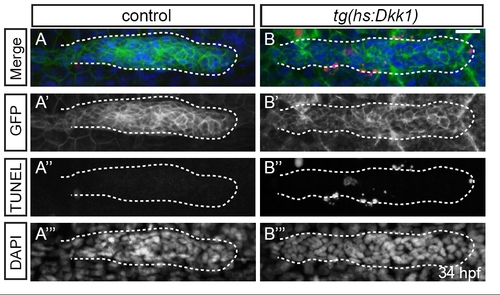
Activation of the Tg(hsp70l:dkk1-GFP) transgene leads to cell death in the primordium. (A-B′′′) Confocal projections of 34 hpf primordia of control embryos (A) or Tg(hsp70l:dkk1-GFP) embryos heat shocked at 28 hpf (B). Cell death was detected by TUNEL (A′′,B′′); primordia were visualized by expression of Tg(-8.0cldnb:lynGFP) transgene (A′,B′) and nuclei were labeled by DAPI (A′′′,B′′′). Although no TUNEL-labeled cells were present in the control primordium (A-A′′′), a number of cells are TUNEL-positive in Tg(hsp70l:dkk1-GFP) embryos heat shocked at 28 hpf (B-B′′′), Scale bar: 20 μm. EXPRESSION / LABELING:
|