- Title
-
Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish
- Authors
- Emoto, Y., Wada, H., Okamoto, H., Kudo, A., and Imai, Y.
- Source
- Full text @ Dev. Biol.
|
Phenotypes of the gir mutant. (A and B) Morphology of the wild type (A) and gir mutant (B) embryos at 36 hpf. The gir mutant embryo exhibits a shortened tail. (C and D) Blood cells in dorsal aorta and posterior cardinal vein at 36 hpf. Circulating blood cells are evident in both dorsal aorta and posterior cardinal vein in wild-type embryos (C), whereas they are absent in the dorsal aorta and have accumulated in the posterior cardinal vein in gir mutant embryos (D). Photographs were taken after the embryos had been cooled on ice to make the circulation slower. (E and F) o-dianisidine staining of blood cells in wild type (E) and gir mutant (F) embryos at 28 hpf. Blood cells are seen in the common cardinal vein and heart in the wild-type embryo (E), whereas these cells have accumulated in the posterior cardinal vein and have failed to circulate in the gir mutant embryo (F). (G and H) Morphology of the wild-type (G) and gir mutant (H) embryos at 80 hpf. The gir mutant shows severe edema. (I and J) Expression of flk1 at 28 hpf. flk1 is expressed in the blood vessels including the common cardinal vein in wild-type embryo (I), whereas its expression is absent in the common cardinal vein of the gir mutant embryo (J). The arrows indicate the common cardinal vein. CCV, common cardinal vein; DA, dorsal aorta; h, heart; N, notochord; PCV, posterior cardinal vein; RBC, red blood cells. EXPRESSION / LABELING:
|
|
Patterning defects in the pectoral fin bud and tailbud in the gir mutant. (A and B) Morphology of the pectoral fin bud in wild type (A) and gir mutant (B) embryos at 36 hpf. The arrowhead indicates the fin bud (A). The gir mutant embryo has no fin bud (B). (C and D) Dorsal views of the wild type (C) and gir mutant (D) embryos at 80 hpf. The arrows indicate pectoral fins. No or small fins were observed in the gir mutant embryo (D). (E–L) Expression of tbx5.1 (E–H), shh (I and J), and ntl (K and L) was examined by in situ hybridization in wild type (E, G, I, and K) and gir mutant (F, H, J, and L) embryos. The expression domain of tbx5.1 has shifted anteriorly in the gir mutant at 20 hpf (F) and 28 hpf (H). The fin bud expression of shh (shown by arrows in I) is absent in the gir mutant at 28 hpf (J). ntl expression is reduced in the tailbud of the gir mutant at the 7-somite stage (L). Genotypes were determined by PCR assay after the photographs had been taken in E, F, K, and L. EXPRESSION / LABELING:
|
|
Molecular isolation of gir. (A) Genetic mapping of gir. In our mapping panel, gir was mapped between Z9891/Z10352 and Z23536 (0.7 cm from Z9891/Z10352 and 1.3 cm from Z23536, n = 150) and at the same position as Z27025. In HS panel, the cyp26a1 gene has been mapped at the same position as Z10352. In the T51 panel, fb81e05, an EST clone of cyp26a1, has been mapped near Z23536. (B) The gir mutant carries a nonsense mutation in the cyp26a1 gene at position 814. The mutated nucleotide is indicated by the arrow. (C) Schematic representation of the Cyp26a1 protein and its truncated form in the gir mutant. The red box represents a heme-binding motif, which is conserved among cytochrome P450s. (D) The cyp26a1 gene is linked to gir. A cyp26a1 fragment was amplified from the wild-type (+/+ or gir/+) and gir mutant embryos and digested with XbaI, which cleaves the mutant cyp26a1 allele, but not the wild-type allele. All gir mutants had only the mutant cyp26a1 allele (no recombinants among 356 meioses), indicating that cyp26a1 is tightly linked to gir. |
|
Morpholino knockdown of cyp26a1 and genetic interaction between gir/cyp26a1 and raldh2. (A, C, and E) Uninjected wild-type control embryos and (B, D, and F) wild-type embryos injected with 1 ng cyp26a1-MO. (A and B) Lateral views of the embryos at 48 hpf. (C and D) Dorsal views of the embryos at 80 hpf. (E and F) o-dianisidine staining of blood cells of the embryos at 28 hpf. Phenotypes of the cyp26a1 morphant are similar to those of the gir mutant. (G, I, and K) Uninjected gir mutants and (H, J, and L) gir mutants injected with 4 ng raldh2-MO. (G and H) Lateral views of the embryos at 80 hpf. (I and J) Dorsal view of the embryos at 80 hpf. (K and L) o-dianisidine staining of blood cells of the embryos at 48 hpf. Genotypes were determined by PCR assay after the photographs had been taken in H, J, and L. |
|
Expression of cyp26a1 and raldh2 is affected in the gir mutant. Expression of cyp26a1 (A–D and G–I), cyp26a1 and krox20 (E and F), and raldh2 (J–O) in the wild type (A, C, E, G, H, J, L, and N) and gir mutant (B, D, F, I, K, M, and O) embryos. (A and B) cyp26a1 expression in the presumptive anterior neural ectoderm and in the tailbud is little affected at bud stage in the gir mutant. Dorsal views are shown. (C and D) At the 5-somite stage, cyp26a1 expression in the rostral presumptive spinal cord is upregulated in the gir mutant. (E and F) At 20 hpf, cyp26a1 expression is strongly upregulated in the rostral spinal cord and in the tailbud of the gir mutant. (G) A transverse section at the level of the rostral spinal cord showing cyp26a1 expression in the wild-type embryo at 20 hpf. N, notochord. (H and I) cyp26a1 expression in the developing branchial arches (indicated by the arrowheads) is perturbed at 28 hpf in the gir mutant. (J–M) raldh2 expression in the presomitic mesoderm and in the somites is reduced in the gir mutant at bud stage (K) and 20 hpf (M). (N and O) At 28 hpf, raldh2 expression in the posterior branchial arch primordium (ba) and in the posterior fin bud- and lateral plate-mesoderm (lp) is disturbed in the gir mutant. Genotypes were determined by PCR assay after the photographs had been taken in A–F and J–M. |
|
Patterning of hindbrain is affected in the gir mutant. Expression of krox20 and myoD (A and B), valentino and myoD (C and D), pax2.1 and krox20 (E and F), gbx2 and krox20 (G and H), eph-b2 (I and J), and otx2 (K and L) in wild type (A, C, E, G, I, and K) and gir mutant (B, D, F, H, J, and L) embryos. (A–D) The distance between the rhombomere 5 (krox20) or 6 (valentino) and the first somite is expanded, and the expression domain of valentino is reduced, in the gir mutant at the 14-somite stage. (E and F) The distance between midbrain–hindbrain boundary (pax2.1) and rhombomere 3 (krox20) is reduced in the gir mutant at the 14-somite stage. (G and H) The distance between rhombomere 1 (gbx2) and rhombomere 3 (krox20) is slightly reduced in the gir mutant at the 10-somite stage. (I and J) The distance between rhombomere 7 and the first somite is expanded in the gir mutant at the 12-somite stage. (K and L) The otx2 expression domain in the midbrain was reduced in the gir mutant at the 10-somite stage. Dorsal views (A, B, G, and H) and lateral views (C–F and I–L) are shown. Genotypes were determined by PCR assay after the photographs had been taken. r, rhombomere; s, somites. EXPRESSION / LABELING:
|
|
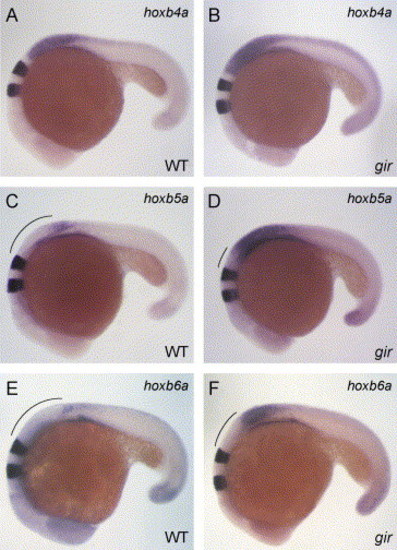
Patterning of the spinal cord is affected in the gir mutant. Expression of hoxb4a and krox20 (A and B), hoxb5a and krox20 (C and D), and hoxb6a and krox20 (E and F) in wild type (A, C, and E) and gir mutant (B, D, and F) embryos at 20 hpf. Expression of these hox genes is upregulated and the expression domains of hoxb5a and hoxb6a are markedly expanded rostrally in the gir mutant. Genotypes were determined by PCR assay after the photographs had been taken. EXPRESSION / LABELING:
|
|
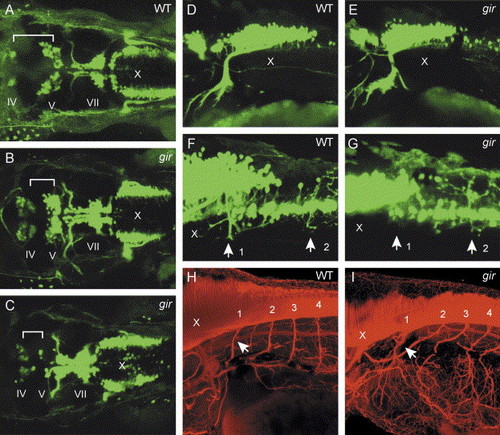
Patterning of motor neurons is affected in the gir mutant. (A–G) Expression of the Isl1-GFP transgene was examined by fluorescence microscopy. (A–E) Dorsal views of hindbrain (A–C) and lateral views of caudal hindbrain and rostral spinal cord (D and E) in wild type (A and D) and gir mutant (B, C, and E) embryos at 48 hpf. The distance between the trochlear (nIV) and trigeminal (nV) nuclei is reduced, and bilateral clusters of the vagal nucleus (nX) are broadened, in the gir mutant (B and C). In some of the gir mutants, the number of neurons in the trigeminal nucleus (nV) is reduced (C). (F and G) Lateral views of caudal hindbrain and rostral spinal cord in wild type (F) and gir mutant (G) embryos at 3 dpf. Dorsal projection of motor neuron axons in the rostralmost spinal cord (labeled by arrow 1) is absent in the gir mutant (G). (H and I) Immunostaining of motor neurons with acetylated α-tubulin antibody in the caudal hindbrain and rostral spinal cord of wild type (H) and gir mutant (I) embryos at 3 dpf. Axons of spinal motor neurons are not significantly affected in the gir mutant (I). Arrows indicate axons from the rostralmost spinal motor neurons. |
Reprinted from Developmental Biology, 278(2), Emoto, Y., Wada, H., Okamoto, H., Kudo, A., and Imai, Y., Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish, 415-427, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.