PUBLICATION
Spliceosomal protein eftud2 mutation leads to p53-dependent apoptosis in zebrafish neural progenitors
- Authors
- Lei, L., Yan, S.Y., Yang, R., Chen, J.Y., Li, Y., Bu, Y., Chang, N., Zhou, Q., Zhu, X., Li, C.Y., Xiong, J.W.
- ID
- ZDB-PUB-161203-16
- Date
- 2017
- Source
- Nucleic acids research 45(6): 3422-3436 (Journal)
- Registered Authors
- Xiong, Jing-Wei
- Keywords
- none
- Datasets
- GEO:GSE78106
- MeSH Terms
-
- Animals
- Apoptosis*
- Brain/abnormalities
- Cloning, Molecular
- Exons
- PubMed
- 27899647 Full text @ Nucleic Acids Res.
Abstract
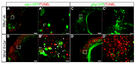
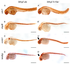
Haploinsufficiency of EFTUD2 (Elongation Factor Tu GTP Binding Domain Containing 2) is linked to human mandibulofacial dysostosis, Guion-Almeida type (MFDGA), but the underlying cellular and molecular mechanisms remain to be addressed. We report here the isolation, cloning and functional analysis of the mutated eftud2 (snu114) in a novel neuronal mutant fn10a in zebrafish. This mutant displayed abnormal brain development with evident neuronal apoptosis while the development of other organs appeared less affected. Positional cloning revealed a nonsense mutation such that the mutant eftud2 mRNA encoded a truncated Eftud2 protein and was subjected to nonsense-mediated decay. Disruption of eftud2 led to increased apoptosis and mitosis of neural progenitors while it had little effect on differentiated neurons. Further RNA-seq and functional analyses revealed a transcriptome-wide RNA splicing deficiency and a large amount of intron-retaining and exon-skipping transcripts, which resulted in inadequate nonsense-mediated RNA decay and activation of the p53 pathway in fn10a mutants. Therefore, our study has established that eftud2 functions in RNA splicing during neural development and provides a suitable zebrafish model for studying the molecular pathology of the neurological disease MFDGA.
Genes / Markers
Expression
Phenotype
Mutations / Transgenics
Human Disease / Model
Sequence Targeting Reagents
Fish
Orthology
Engineered Foreign Genes
Mapping