Fig. 2.
- ID
- ZDB-FIG-221226-122
- Publication
- Do et al., 2022 - Hypoxia deactivates epigenetic feedbacks via enzyme-derived clicking proteolysis-targeting chimeras
- Other Figures
- All Figure Page
- Back to All Figure Page
|
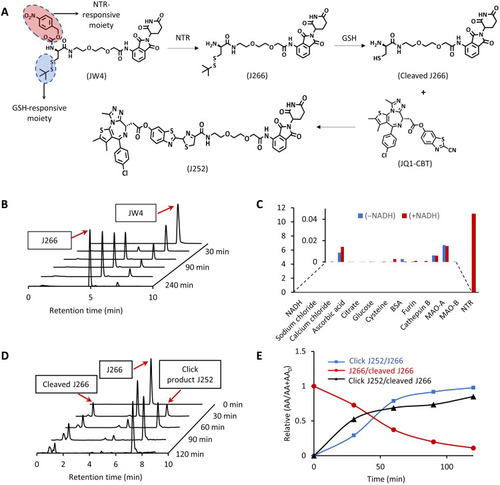
A) Chemical structure of GSH-responsive CRBN ligand (J266); GSH- and NTR-responsive CRBN ligand (JW4); cleaved J266; CBT-linked BRD4-targeting ligand (JQ1-CBT); click-induced BRD4 degrader (J252). (B) LC-MS spectra of NTR uncaging JW4 (10 μM) to form J266 at different time points in NTR (40 μg/ml) dissolved in phosphate-buffered saline (PBS; 10 mM) (pH 7.4). (C) Selectivity of JW4 toward a broad range of biological and chemical agents with or without NADH. (D) Time-dependent LC-MS spectra of click J252 formation under reducing reagent [tris(2-carboxyethyl)phosphine (TCEP)] that cleave the J266 to induce cleaved J266 before the click reaction with JQ1-CBT in buffer solution. (E) Ratio of peak areas of click J252 and cleaved J266 to J266 over multiple time points in (D). |