Fig. 1
- ID
- ZDB-FIG-200406-19
- Publication
- Mikalsen et al., 2020 - Phylogeny of teleost connexins reveals highly inconsistent intra- and interspecies use of nomenclature and misassemblies in recent teleost chromosome assemblies
- Other Figures
- All Figure Page
- Back to All Figure Page
|
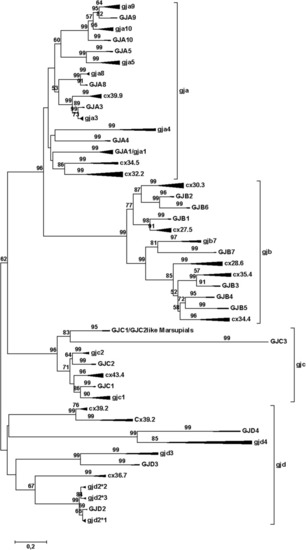
Phylogenetic tree for the gap junction protein (connexin) gene family. The mammalian branches are indicated by upper case letters; teleost branches are indicated by lower case letters. The width of the triangles indicates the number of taxa included in the branch, and the length of the triangles indicates the sequence variation within the branch. The tree was made by the Minimum Evolution method, using amino acids (354 amino acid sequences with 201 positions in the final dataset) and the Dayhoff substitution matrix. The bootstrap values (500 replicates) > 50% are shown next to the branches. To avoid disruptive long-branch attraction, some sequences were excluded (see text). This model gives results that are quite close to the majority of results as summed up in Suppl. Table |