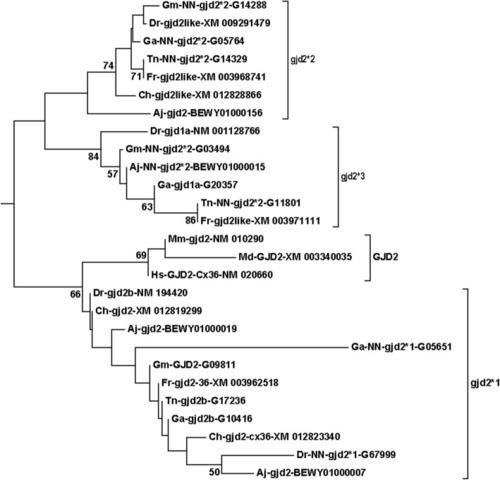
Fig. 4
- ID
- ZDB-FIG-200406-22
- Publication
- Mikalsen et al., 2020 - Phylogeny of teleost connexins reveals highly inconsistent intra- and interspecies use of nomenclature and misassemblies in recent teleost chromosome assemblies
- Other Figures
- All Figure Page
- Back to All Figure Page
|
The |