- Title
-
Zebrafish Mbd5 binds to RNA m5C and regulates histone deubiquitylation and gene expression in development metabolism and behavior
- Authors
- Guo, J., Zou, Z., Dou, X., Zhao, X., Wang, Y., Wei, L., Pi, Y., Wang, Y., He, C., Guo, S.
- Source
- Full text @ Nucleic Acids Res.
|
|
|
Germ-line knockouts of |
|
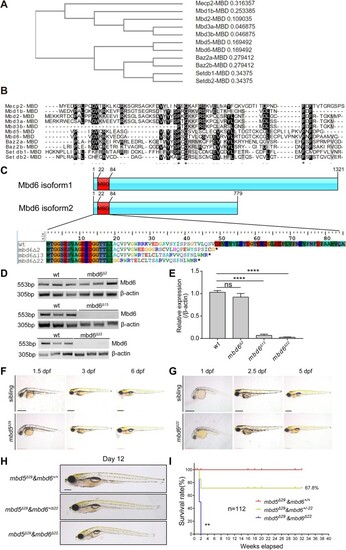
CRISPR/Cas9-mediated germ-line disruption of Mbd6, a MBD protein closely related to Mbd5, reveals redundant function in larval growth and physiology. ( |
|
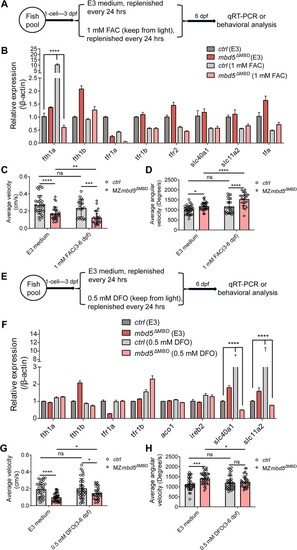
Alterations in iron metabolism contribute to behavioral abnormalities in the |
|
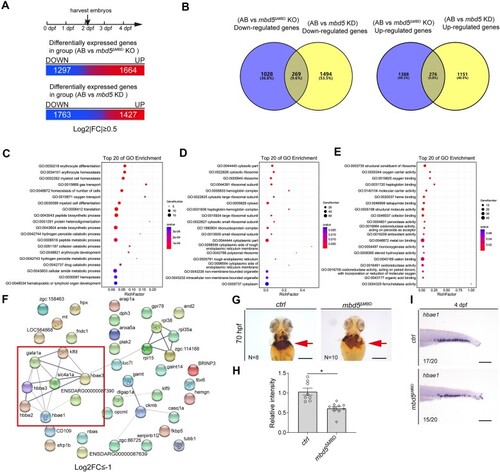
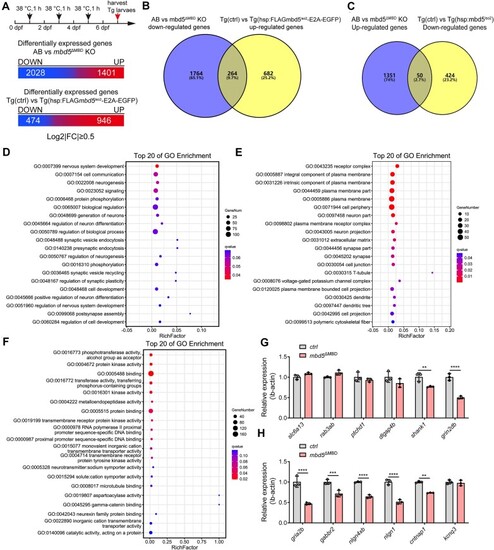
RNA-seq analyses uncover significantly down-regulated biological processes and pathways and highlight deficiency of erythroid differentiation common in both |
|
|
|
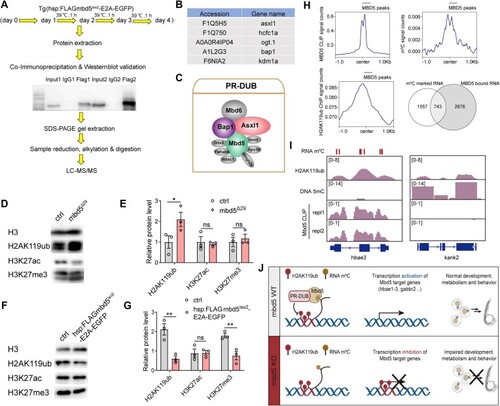
Mbd5 preferentially binds to m5C-modified mRNAs and regulates target transcript abundance. ( |
|
Mbd5 interacts with PR-DUB and is necessary and sufficient to promote deubiquitylation. ( |