- Title
-
wnt10a is required for zebrafish median fin fold maintenance and adult unpaired fin metamorphosis
- Authors
- Benard, E.L., Küçükaylak, I., Hatzold, J., Berendes, K.U.W., Carney, T.J., Beleggia, F., Hammerschmidt, M.
- Source
- Full text @ Dev. Dyn.
|
Characterization of fin defects in unm_t30922 mutants. (A) unm_t30922 mutant embryos are initially identical to their siblings at 35 hpf and form regular median fin folds. (B) Both the minor and major lobes of the MFF have started to collapse at 48 hpf. (C-D‴) At 6 dpf, the MFF of the sibling has extended (C, D, D′) while most of the mutant MFF has collapsed (C, D″, D‴) apart from small remnants visible in the minor lobe (arrowheads in D″) and large parts of the tail fin fold (D‴). (E) Alcian blue stainings reveal that at 6 dpf, the pectoral fins of siblings and mutants are indistinguishable and have regular cleithrum (CL), scapulocoracoid (SC), endoskeletal disc (ED) cartilages, and a regular fin fold (FF). (F, G) Alizarin red stainings of mineralized bones (F) and images of live fish (G) reveal that at the onset of metamorphosis (15 dpf, SL 6 mm) wnt10a mutants only have remnants of caudal fins with disorganized lepidotrichia (F, G), while the minor and all other parts of the major MFF lobes have disappeared (G). Arrows in the wild-type fish point to the forming unpaired anal (af), dorsal (df), and caudal (cf) fin. (H) As juveniles (26 dpf, SL 11 mm; osterix:nuGFP transgenic18 to label bone-forming cells) the mutants lack the unpaired dorsal (df) and anal (af) fin, while the unpaired caudal fin (cf) is strongly reduced, containing only few lepidotrichia. In comparison, the paired pectoral fins (pecf) and pelvic fins (pelf) are little affected. In addition, scales are formed as in their siblings (enlarged inserts with dashed lines). Arrowheads indicate collapsed fin folds and remnants of adult fins. MFF, median fin fold; SL, standard length. |
|
unm_t30922t30922 mutants carry a homozygous splice acceptor site mutation within the wnt10a gene, representing a likely wnt10a amorph. (A) Meiotic mapping placed the MFF collapse-causing mutation into a 0.429 cM interval of chromosome 9 between SSLP markers z25735 (2/1400 recombinants/meioses) and NZ79 (4/1400 recombinants/meioses). (B, C) Whole-genome sequencing (WGS) of DNA from 20 pooled mutant embryos revealed a region of homozygosity exclusively on chromosome 9 (B), which was approximately 4 Mb large (indicated by red lines in (C)) and contains a single point mutation (A>G) in the splice acceptor site before exon 4 of wnt10a (C). (D) Sequencing of cDNA from mutant embryos revealed three mutant splice isoforms, resulting from the usage of cryptic splice acceptor sites within intron 3/4 or exon 4 and leading to C-terminally truncated Wnt10a proteins lacking exon 4-encoded amino acid residues 288–442. (E) Quantitative real-time PCR (qRT-PCR) of whole embryos shows reduced amounts of wnt10a transcripts in unm_t30922t30922 mutants at 2 and 5 dpf. |
|
unm_t30922t30922 mutants display impaired tooth development. (A) At 6 dpf, the wnt10a siblings have alizarin red-stained 3V1, 4V1, and 5V1 teeth on either bilateral ceratobranchial 5 arch (cb5), while the unm_t30922t30922 mutant either has one tooth or no teeth on either side of an otherwise normally formed cb5. The cleithra (CL) of the mutant is indistinguishable from those of the siblings. (B) At 35 dpf (SL 13 mm), dissected cb5 of siblings have fully formed alizarin red-stained mineralized teeth (only 1V, 2V, 3V, 4V, and 5V are visible in this view) while the unm_t30922t30922 mutants have no teeth and an underdeveloped cb5. Arrows indicate regular teeth, arrowheads compromised or absent teeth. Scale bars = 100 μm. (C) Dorsal view of wild-type embryo at 56 hpf, after wnt10a in situ hybridization shows that wnt10a is expressed in the tooth germs (indicated by arrows). (D) Transverse cryosection of 56 hpf wild-type embryo after wnt10a in situ hybridization and p63 immunostaining, revealing strong wnt10a expression in the dental mesenchyme (dm; white arrow) and weak wnt10a expression in the surrounding p63-positive dental epithelium (de; white arrow). (E) Dorsal view of Tg(6xTCF:eGFP) embryo at 56 hpf after transgene-encoded GFP in situ hybridization, revealing the reception of canonical Wnt signals in the tooth germs (arrows). (F) Transverse cryosection of Tg(6xTCF:eGFP) embryo at 56 hpf after transgene-encoded GFP in situ hybridization and p63 immunostaining, revealing strong canonical Wnt signal reception in the dental mesenchyme (dm; white arrow) and weak wnt10a expression in the surrounding p63-positive dental epithelium (de; white arrow). (G, H) Durcupan transverse sections of in situ hybridized unm_t30922t30922 sibling and mutant embryos at 56 hpf. eda is strongly expressed in the dental mesenchyme (dm, arrow) and weakly expressed in the dental epithelium (de, arrow) of both the unm_t30922t30922 sibling and mutant tooth germ (G). fgf4 is strongly expressed in tooth germ (arrow) of the unm_t30922t30922 sibling but not in the tooth germ of the unm_t30922t30922 mutant (arrowhead) (H). (I) Transverse cryosection of unm_t30922t30922 sibling at 56 hpf after fgf4 in situ hybridization and p63 immunostaining, revealing strong fgf4 expression in the p63-negative dental mesenchyme. (J) Dorsal views and transverse Durcupan sections of in situ hybridized unm_t30922t30922 sibling and mutant embryos at 56 hpf revealing strong dlx2b expression in the dental mesenchyme (dm) and dental epithelium (de) of the sibling (arrows), but an almost complete down-regulation in the mutant (arrowhead). de, dental epithelium; dm, dental mesenchyme; n, notochord; pecf, pectoral fin; y, yolk sac. |
|
The fin defects of unm_t30922t30922 mutants can be phenocopied by loss and rescued by gain of Wnt10a function. (A, B) Morpholino knockdown of wnt10a (A) or inhibition of canonical Wnt signaling via temporally controlled transgenic expression of dkk1 from 24 or 48 hpf onwards (B) both phenocopy the median fin fold collapse observed in the unm_t30922t30922 mutant embryos at 3 dpf. (C) Transgene-encoded wild-type wnt10a can fully rescue MFF collapse of unm_t30922t30922 mutants at 3 dpf when expressed from 24 hpf onwards, but can only rescue partially when expressed from 48 hpf onwards. (D) At 14 dpf (SL 5 mm), unm_t30922t30922 mutants lack most of their MFFs with the exception of the caudal fin. Similar to the effects at 3 dpf, mutants with transgenic expression of wild-type wnt10a from 24 to 144 hpf have regular MFFs, while mutants with transgenic expression of wild-type wnt10a from 48 to 144 hpf lack parts of their MFFs. (E) Despite the complete or partial MFF rescue at 3 and 14 dpf, mutants with transgene-driven wild-type wnt10a expression from 24 or 48 to 144 hpf have failed to develop complete adult fins at 31 dpf (SL 9 mm), resembling unm_t30922t30922 mutants without forced wild-type wnt10a expression. Arrowheads indicate collapsed fin folds and remnants of adult fins. MFF, median fin fold; SL, standard length. |
|
wnt10a is expressed in fin mesenchymal cells. wnt10a in situ hybridization of wild-type embryos. (A) At 28 hpf, wnt10a is expressed in the fin mesenchymal cells, as the cells start to migrate into the dermal space of the MFF (arrows in A', showing a magnified view of region framed in (A)). (B) At 56 hpf, wnt10a is expressed in fin mesenchymal cells throughout the entire dermal space of the MFF (arrow in B′, showing a transverse section at the position indicated by dashed line in (B)). (C) At the onset of metamorphosis at 13 dpf (SL 5 mm), wnt10a is strongly expressed in the dermal space of the caudal fin where lepidotrichia start to form (arrows in (C′), showing a transverse section at the position indicated by dashed line in (C)). MFF, median fin fold; SL, standard length. |
|
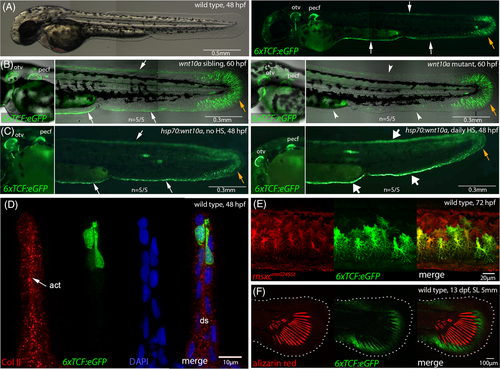
Canonical Wnt signals are received by distal-most epidermal cells and fin mesenchymal cells of the embryonic MFF. Fluorescent images of live Tg(6xTCF:eGFP) transgenics, with cells receiving canonical Wnt signals in green. (A) At 48 hpf, the responder transgene is expressed in distal-most cells of the MFF (white arrows) and in fin mesenchymal cells (orange arrow) of the caudal fin fold of wild-type embryo. (B) Compared to wild-type sibling (left panel), wnt10a mutant embryo of 60 hpf (right panel) shows reduced Tg(6xTCF:eGFP) expression in the minor and major lobes of the MFF (indicated by arrowheads), whereas expression in the caudal fin fold (indicated by orange arrow) and in the AER of the pectoral fin is comparably unaltered. (C) Compared to non-heat-shocked control (left panel), transgenic embryo with heat-shock-induced global wnt10a overexpression from 24 to 48 hpf (right panel) shows increased Tg(6xTCF:eGFP) expression in the minor and major lobes of the MFF (indicated by thick arrows), whereas expression in the caudal fin fold (indicated by orange arrow) and the AER of the pectoral fin is unaltered. (D) Transverse cryosection approximately 0.5 mm anterior of the caudal tip of the tail of 6xTCF:eGFP 48 hpf embryo, stained with antibodies against GFP and collagen II (labeling actinotrichia, red). The Wnt responder is expressed in distal-most positions of the MFF. (E) 6xTCF:eGFP, GBT0245 (msxc; marker of fin mesenchymal cells in red55) double transgenic embryo revealing expression of the Wnt responder in fin mesenchymal cells of the ventral MFF at 3 dpf. (F) At the onset of metamorphosis (13 dpf, SL 5 mm), the Wnt responder is expressed distal of forming lepidotrichia (stained with alizarin red) of the caudal fin. act, actinotrichia; AER, apical ectodermal ridge; ds, dermal space; MFF, median fin fold; otv, otic vesicle; pecf, pectoral fin; SL, standard length. |
|
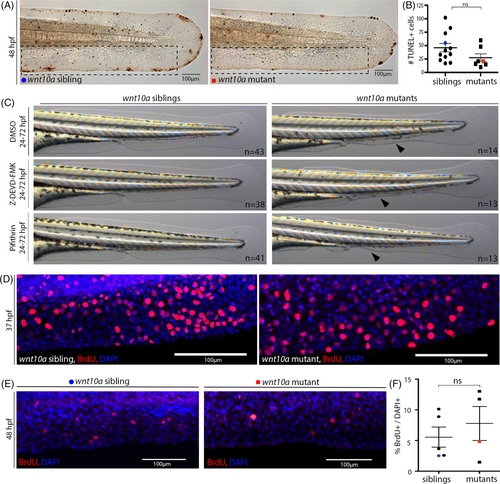
MFF collapse in wnt10a mutant does not involve increased cell death or impaired cell proliferation. (A) TUNEL staining shows no major differences in the number of dying cells in the MFF of wnt10a mutant embryos at 48 hpf. (B) Quantification of the number of TUNEL-positive cells per embryo within region framed in (A), indicating no statistically significant increase in mutants compared to siblings. The blue dot and red square indicate values for embryos shown in (A). (C) Treatment from 24 to 72 hpf with the Caspase-3 inhibitor Z-DEVD-FMK (80 μM), the p53 inhibitor Pifithrin-α (0.6 μM), and DMSO (control) does not prevent the MFF collapse of wnt10a mutants at 72 hpf. (D, E) BrdU incorporation (in red) indicates no obvious differences in cell proliferation within the ventral MFF at 37 hpf, when MFF collapse commences (D), or at 48 hpf ((E), nuclei stained with DAPI in blue), when MFF is in progress (E). (F) Percentages of BrdU-positive cells in ventral MFF within the region shown in (E), indicating no statistically significant decrease in mutants compared to siblings. The blue dot and red square indicate values for embryos shown in (E). MFF, median fin fold. |
|
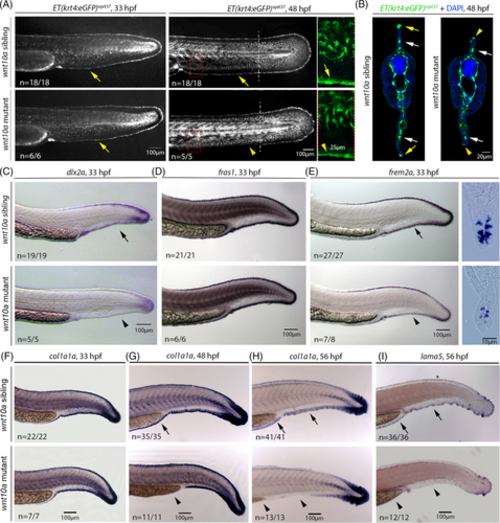
MFF collapse in wnt10a mutant MFF involves the down-regulation of dlx2a and crucial ECM protein-encoding genes in distal MFF cells. (A) At 33 hpf (left panels), before MFF collapse becomes morphologically visible, wnt10a mutant displays unaltered expression of the Et(krt4:eGFP)sqet37 transgene both in fin mesenchymal cells and in distal-most epidermal cells of the MFF (indicated by yellow arrows). At 48 hpf (middle panels and right panels for enlarged views of regions framed in middle panels), when MFF collapse is obvious and in progress, fin mesenchymal cells of the mutant still show normal Et(krt4:eGFP)sqet37 expression, whereas transgene expression in distal epidermal cells is reduced (indicated by yellow arrowheads) in comparison to wild-type sibling (indicated by yellow arrow). (B) Transverse sections of 48 hpf Et(krt4:eGFP)sqet37 transgenics at the location indicated in middle panels of (A) by dashed lines, indicating the normal positioning and transgene expression in fin mesenchymal cells (white arrows), but the reduced transgene expression in distal epidermal cells both in the collapsing dorsal and ventral MFF of the mutant (yellow arrowheads) compared to its sibling (yellow arrows). (C–E) In situ hybridization indicates that at 33 hpf, before MFF collapse becomes morphologically visible, the expression of the distal marker gene dlx2a is strongly down-regulated in distal-most epidermal cells of the ventral MFF (C). Expression of fras1, a marker for distal-most cleft cells,65 appears unaltered in mutants compared to their wild-type siblings (D), whereas expression of frem2a, a marker for distal ridge cells,65 is reduced, as also obvious in transverse Durcupan sections (E). (F–I) In situ hybridization indicates that at 33 hpf (left panels), before MFF collapse becomes morphologically visible, expression of col1a1a, encoding an essential component of actinotrichia, is normally expressed in distal-most epidermal cells of the MFF of wnt10a mutants (F), while expression levels become progressively reduced at 48 hpf ((G); most obvious in minor lobe) and at 56 hpf (H), when fin collapse is in progress. A similar progressive reduction of expression in the MFF of wnt10a mutant is observed for lama5, encoding an essential component of the MFF basement membrane ((I) for 56 hpf; earlier stages not shown). Arrows indicate regular expression; arrowheads indicate less or non-detectable expression. ECM, extracellular matrix; MFF, median fin fold. |
|
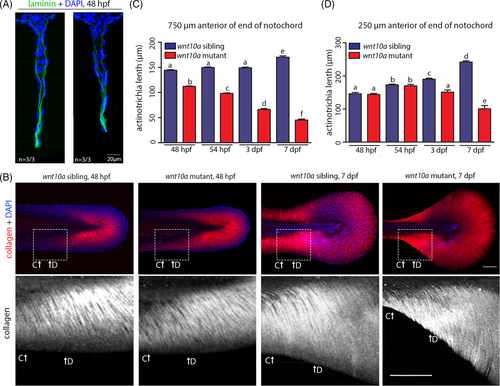
wnt10a mutants display a progressive shrinkage of MFF actinotrichia. (A) Immunofluorescence of Laminin (in green, nuclei stained with DAPI in blue), transverse sections of tail region approximately 750 μm anterior of the tip of the tail of embryos at 48 hpf. Laminin shows indistinguishable protein levels and basement membrane localization in wnt10a sibling and mutant ventral MFFs. (B) Actinotrichia, visualized via Col II immunofluorescence (in red in upper row and white in lower row; nuclear DNA stained with DAPI in blue) in the ventral MFF of wnt10a mutants and siblings at 48 hpf and 7 dpf. Lower row shows magnified views of regions framed in upper row. (C, D) Graphical illustration of lengths of actinotrichia at two corresponding locations indicated in (B) in the major lobe of the ventral MFF in wnt10a siblings and mutants at 48 hpf, 54 hpf, 3 dpf, and 7 dpf. MFF actinotrichia continue to grow in the sibling, whereas they become progressively shorter in the mutant. |
|
Actinotrichia of the MFF of wnt10a mutants do not extend properly into the cleft of epidermal cleft cells. All images show TEM micrographs of transverse sections through the ventral MFFs of wnt10a siblings and mutants of 48 hpf, proximately 750 μm anterior of the tip of the caudal fin. (A) In intermediate proximodistal positions, the ventral MFF mutants display regular ultrastructure, including the basement membrane between epidermal cells and the dermal space (arrowheads), basement membrane–dermal anchorage and actinotrichia. (B) In distal-most positions of the ventral MFF, cleft cells of wnt10a mutants display a slightly widened cleft (indicated by blue and red brackets for sibling and mutant, respectively). In addition, in contrast to siblings, in mutants, actinotrichia do not reach the distal tip of the cleft (distal end of actinotrichia in sibling and mutant indicated by blue and red arrowheads, respectively). Furthermore, siblings show actinotrichia extending into the interior of cleft cells, constituting “fibripositor”-like structures71 (indicated by yellow arrow in sibling), whereas mutants do not. Blue, yellow, and red dashed rectangles of overview images frame regions shown in magnified views to their right. (C, D) Graphical illustration of quantification demonstrating that wnt10a mutant embryos do not have significantly shorter ventral MFFs compared to the siblings (C), but a statistically significant increase in the distance between actinotrichia and the distal tip of the cleft of cleft cells (D). The blue dots and red squares in (C, D) indicate values for the respective sibling and mutant shown in (B). (E) At the level of ridge cells, actinotrichia of siblings and mutants display comparable width and banded structure. MFF, median fin fold. |