- Title
-
Using single-molecule fluorescence in situ hybridization and immunohistochemistry to count RNA molecules in single cells in zebrafish embryos
- Authors
- Keseroglu, K., Zinani, O.Q.H., Özbudak, E.M.
- Source
- Full text @ STAR Protoc
|
|
|
Critical experimental steps (A and B) (A) Image of dissecting microscope and (B) its mirror position. (C) A representative image of fixed embryo with transparent tissue. (D) Schematic representation how tubes should be seen after air dry. (E) Pipette tip position (horizontal) while adding a solution to prevent damage of embryos. (F–H) (F) Horizontal, (G) tilted, and (H) vertical positions of tube in a box during shaking on the shaker or incubation in the oven. |
|
Flat-mounted sample preparation (A) Cut PSM from embryo using microsurgery knife. (B) Transfer tissue from petri dish into a drop of ProLong Gold antifade reagent on a glass slide. (C) Flattened tissue in the reagent. (D) Apply nail polish and fix the sample with a coverslip. (E) Dorsoventral view of a flat-mounted PSM tissue with couple of somites. |
|
Microscopy settings for imaging (A and B) Microscopy settings for (A) DAPI-TRITC-CY5 and (B) FITC channels. Use separate optical configurations for DAPI-TRITC-CY5 and FITC because of different integrate levels. (C) “Scan Large Image” window, where left-top-right-bottom limits of the tissue and number of tiles in XY dimensions are detected. (D) Settings for Z layer in the relative symmetric mode. Z-step size should be 0.27 μm. (E) Window shows adding custom XY multipoints. (F) Positions of XY multipoints that covers the tissue. Remove unnecessary multipoints in case corner tiles are free of tissue. (G) Multichannel panel, which includes DAPI-TRITC-CY5 and FITC optical configurations. |
|
Surface creation and masking image (A) Stitch ND2 raw image. One of the (B) Converted stitched file to Imaris format. (C) Select tissue format in every 5 slices using manual surface creation. (D and E) Created surface to mask DAPI (nucleus) and FITC (membrane) channels. (F) Use “Mask All” in the “Tab Edit”. Set voxel intensity outside surface to zero “0”. (G) Masked tissue. (H and I) (H) Use “free rotate” window to make image anterior-posterior view (I) from left to right. |
|
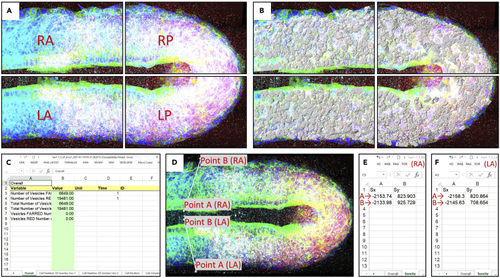
Cell segmentation for analysis and finding PSM-somite border (A) Split image into four images using “Crop 3D”. One of the (B) Segmented cells are seen in the Right-Anterior, Right-Posterior, Left-Anterior, and Left-Posterior of the tissue. (C) An example of XLS file, which includes statistics of “Cell Number of Vesicles”, “Cell Position”, and “Cell Volume”. (D) Measurement points for anterior PSM border. (E and F) Add X and Y data of the points in the “Somite” sheet of the (E) left-anterior and (F) right-anterior XLS file. |
|
Measuring angle of expression stripes along the posterior-anterior axis (A–C) Measuring angle of expression stripes along the posterior-anterior axis and linear fitted equation for angle data (A), (B), and (C) show different wild-type samples as an example for measuring angle and distance of stripe to tailbud. Scale bar is 100 µm. (D) Plot “Angle vs Distance (μm)” graph to fit an equation, where the slope and Y-intersected data provide “ |
|
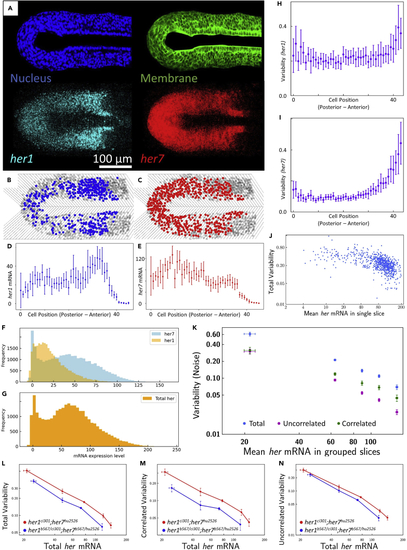
Example outcomes of images, RNA and variability (noise) distributions (A–N) (A) Masked nucleus (blue) and membrane (green) frames, and 3D projection of |
|
Comparison between “good” and “bad” fixed embryos |