- Title
-
Zebrafish Ski7 tunes RNA levels during the oocyte-to-embryo transition
- Authors
- Cabrera-Quio, L.E., Schleiffer, A., Mechtler, K., Pauli, A.
- Source
- Full text @ PLoS Genet.
|
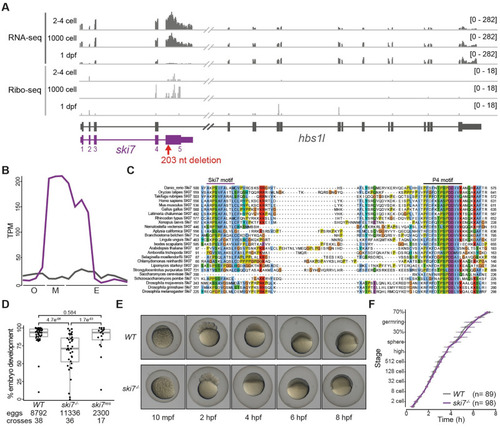
(A) Expression profile of the |
|
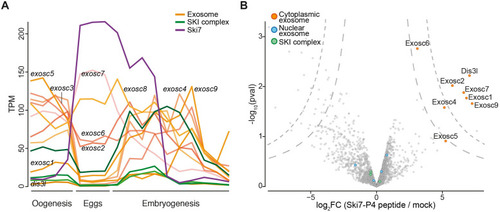
(A) mRNA expression levels of |
|
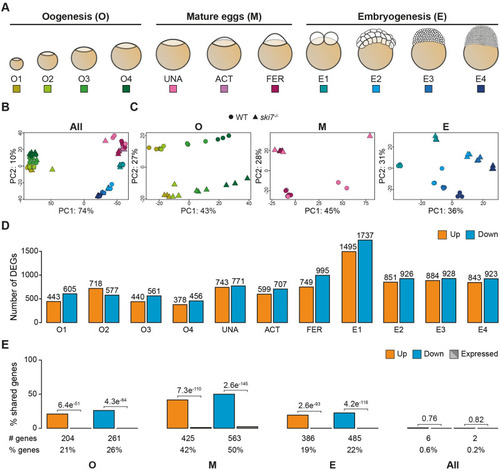
(A) Schematic representation of the stages used for RNA-seq during three consecutive developmental periods: oogenesis, mature eggs and embryogenesis. (B, C) Principal Component Analyses (PCA) of all time points (B) or individual periods (C) used for the polyA+ RNA-seq comparison of |
|
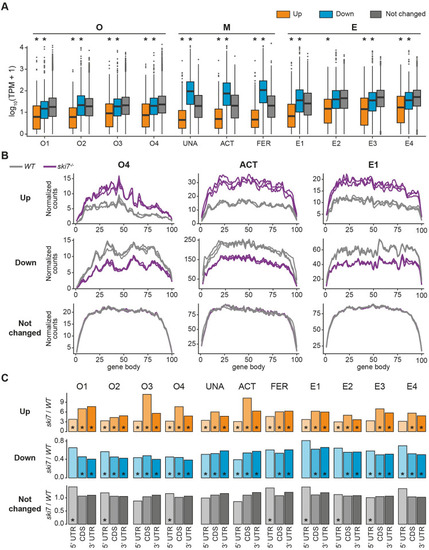
(A) Genes up-regulated in the absence of Ski7 show low expression levels in wild type. Expression levels of DEGs in wild type for every stage of the time course, as measured by transcripts per million (TPM). * indicates p-value < 0.01 from Wilcoxon test in comparison to unchanged genes. (B) Metagene profiles of up-regulated (top), down-regulated (middle), and unchanged genes (bottom) of wild type and PHENOTYPE:
|
|
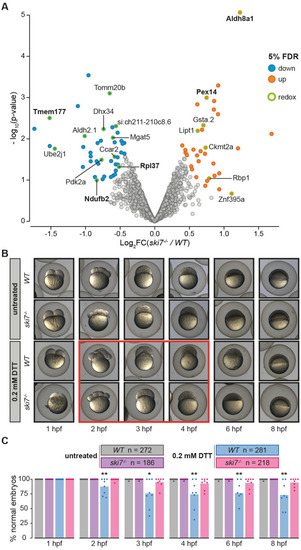
(A) Volcano plot of proteins identified by tandem mass tag mass spectrometry (TMT-MS) from wild-type and PHENOTYPE:
|