- Title
-
The zebrafish Kupffer's vesicle as a model system for the molecular mechanisms by which the lack of Polycystin-2 leads to stimulation of CFTR
- Authors
- Roxo-Rosa, M., Jacinto, R., Sampaio, P., Lopes, S.S.
- Source
- Full text @ Biol. Open
|
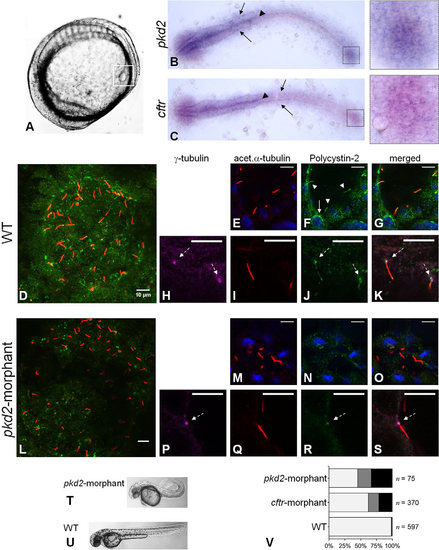
Polycystin-2 and CFTR expression. (A) Localization of KV (squared region) in the body of a 10–11s.s. zebrafish embryo. (B,C) RNA in situ hybridizations for pkd2 (B) and cftr (C) in 10–11s.s. WT embryos. Both pkd2 and cftr transcripts are detected in the KV region (right squares), neural floorplate (arrow heads), brain and pronephric ducts primordia (arrows). (D-S) Confocal images for the immunolocalization of Polycystin-2 in KV cells at the 10–11s.s. In WT embryos (D-K), Polycystin-2 is detected clustered near the nuclei (white arrow in F), along cilia (white arrow heads in F) and at the basal body (dashed arrows in H,J and K). In pkd2-morphants (L-S), the Polycystin-2 signal is markedly reduced and, although still detected at the basal body (dashed arrow in P,R and S), it is no longer detected along cilia. (D,L) maximal intensity z-stack projection; (E-K;M-S) z-section. Polycystin-2 (green), acetylated α-tubulin (red), γ-tubulin, (purple), nuclei (blue). Scale bars: 10µm. (T,U) Lateral view of pkd2-morphant (T) and WT (U) larvae at 72hpf. (V) Heart position defects: pkd2-morphants – 33% right-sided, 21% central; cftr-morphants – 21% right-sided, 17% central; and WT siblings – 0.7% right-sided, 1.0% central. Left-sided (light grey), central (dark grey) and right-sided hearts (black). n, number of scored embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
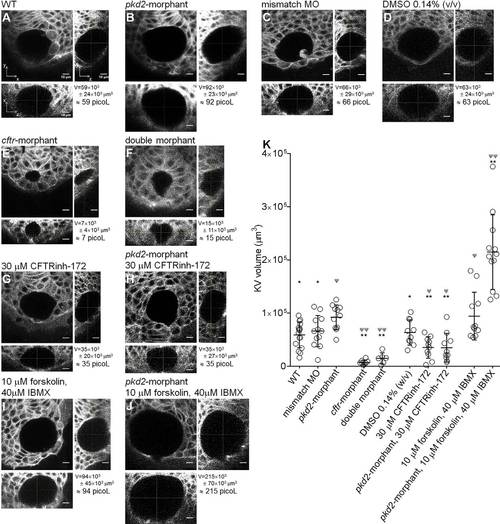
KV volume. (A-J) Confocal live-microscopy scans of the whole KV of 10–11s.s. ras:GFP transgenic embryos. The middle focal plane along the xy axis and the respective orthogonal views (along xz and yz axes) are shown for the most representative WT (A), pkd2-morphant (B), pkd2-mismatch MO (C), 0.14% (v/v) DMSO-treated WT (D), cftr-morphant (E), double-morphant (F), 30µM CFTRinh-172-treated WT (G) and pkd2-morphant (H), and 10µM forskolin+40µM IBMX-treated WT (I) and pkd2-morphant (J) embryos. C is a control for B. D is a control for G,H,I and J. KVvolume is indicated in µm3 and in picol. Scale bars: 10µm. (K) Estimated KV volumes (µm3) for WT (n=16), pkd2-mismatch MO (n=12), pkd2-morphant (n=11), cftr-morphant (n=8), double-morphant (n=6), 0.14% (v/v) DMSO-treated WT (n=10), 30µM CFTRinh-172-treated WT (n=10) and pkd2-morphant (n=10), 10µM forskolin+40µM IBMX-treated WT (n=11) and pkd2-morphant (n=12) embryos. Mean±s.d.; ΨPd0.05 and ΨψP<0.0001, significantly different from WT; *P<0.05 and **P<0.0001, significantly different from pkd2-morphants. EXPRESSION / LABELING:
PHENOTYPE:
|
|
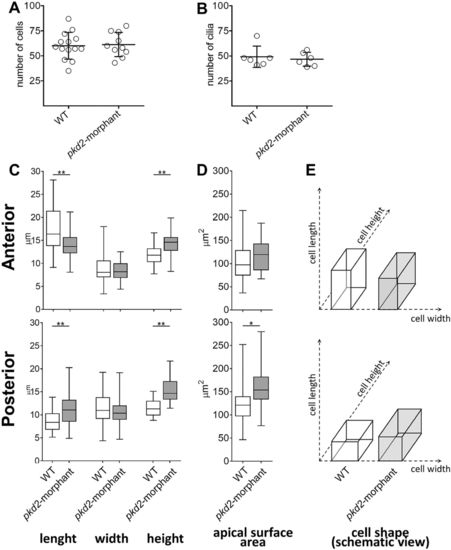
KV-lining cells. (A) Number of cells counted in the whole KV live-microscopy scans of 10–11s.s. WT (n=14) and pkd2-morphant (n=10) embryos. (B) Number of cilia counted in WT (n=6) and pkd2-morphant (n=6) embryos immunodetected for acetylated α-tubulin. (C) Cellular length, width and height of WT (n=6) and pkd2-morphant (n=11) embryos immunodetected for actin cytoskeleton. The box plot and the respective max and min values are indicated. *P<0.01 and **P<0.0001, significantly different from WT. (D) Estimated apical surface area of KV-lining cells. (E) Schematic representation of cell shape of WT and pkd2-morphant KVs. PHENOTYPE:
|
|
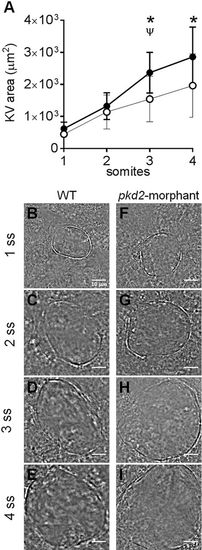
KV inflation live-dynamics. (A) KV luminal area of pkd2-morphants and WT embryos measured along development from 1 to 4 s.s. For each time point mean ±s.d. are indicated. Number of tested embryos: WT, n=11; pkd2-morphant, n=8. ψP<0.05, significantly different from the previous time point in pkd2-morphants; *P<0.05, significantly different from WT at the corresponding time point. (B-I) Bright field images captured for the same embryo along development are shown for the most representative WT (B-E) and pkd2-morphant (F-I). Scale bars: 10μm. PHENOTYPE:
|
|
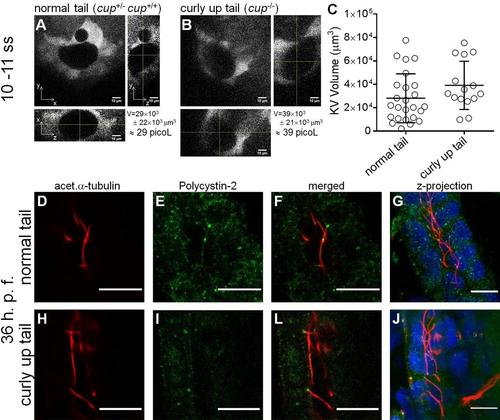
cup mutant KV volumes. Whole KV live-microscopy scans of 10–11 s.s. embryos from cup+/- ; foxj1a:GFP parents. The middle focal plan along the XY axis and the respective orthogonal views (along XZ and YZ axes) are shown for the most representative normal tail (cup+/-; cup+/+) (A), and curly-up tail (cup-/-) (B) embryos. KVvolume is indicated in µm3 and in picoL. Scale bars: 10 µm. (C) Estimated KV volumes (µm3) for normal tail (cup+/-; cup+/+) (n = 23) and curly-up tail (cup-/-) (n = 15) embryos. Average values and the respective s.d. are indicated. EXPRESSION / LABELING:
PHENOTYPE:
|
|
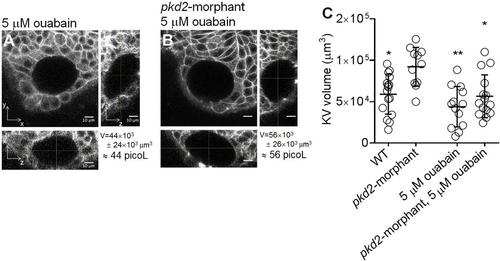
Ouabain effect on KV volume. Live-scans of the whole KV of 10–11 s.s. ras:GFP transgenic embryos. The middle focal plan along the XY axis and the respective orthogonal views (along XZ and YZ axes) are shown for the most representative 5 µM ouabain treated WT (A) and pkd2-morphant (B) embryos. KVvolume is indicated in µm3 and in picoL. Scale bars: 10 µm. (C) Estimated KV volume (µm3) for WT (n = 16), pkd2-mismatch MO (n = 12), 5 µM ouabain treated WT (n = 13) and pkd2-morphant (n = 15) embryos. Average values and the respective s.d. are indicated. * and **, significantly different from pkd2-morphants (p < 0.05 and p < 0.0001, respectively). EXPRESSION / LABELING:
PHENOTYPE:
|