- Title
-
Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling
- Authors
- Tyurina, O.V., Guner, B., Popova, E., Feng, J., Schier, A.F., Kohtz, J.D., and Karlstrom, R.O.
- Source
- Full text @ Dev. Biol.
|
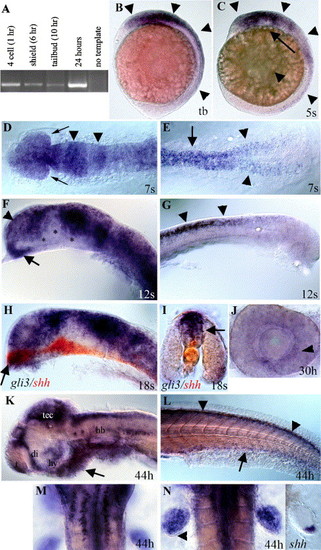
Developmental expression of zebrafish gli3. (A) RT–PCR analysis using gli3-specific primers shows that maternal gli3 mRNA is present at the four-cell stage (prior to zygotic transcription), as well as at shield, tail bud, and 24-h stages, after zygotic transcription has begun. (B) Tail bud stage (10 h). gli3 is first expressed in neural ectoderm (arrowheads) with expression most intense in the brain. (C) Five-somite stage (12 h). During early somitogenesis, gli3 is expressed throughout the brain and spinal cord (arrowheads) with expression being lost in more ventral neural tissue (arrow). (D, E) Seven-somite stage (13 h). Dorsal view. (D) gli3 is expressed throughout the embryonic brain and developing eyes (arrows) with more intense stripes of expression in the presumptive midbrain and midbrain/hindbrain border (arrowheads). (E) In the trunk, gli3 is expressed in the spinal cord (arrow) and in the lateral neural plate prior to neural tube formation (arrowheads). (F, G) Twelve-somite stage (15 h). (F) At 15 h, gli3 is expressed throughout the telencephalon (arrowhead) and anterior diencephalon (arrow) in the forebrain. (G) At 15 h, gli3 is strongly expressed in the spinal cord and is restricted to the dorsal neural tube (arrows). (H, I) Eighteen-somite stage (17 h). (H) Dorsal expression of gli3 (purple/blue) is largely complementary to the more ventral expression of shh (red) in the brain. However, in the most anterior diencephalon (arrow in H), gli3 expression overlaps with shh expression. (I) Cross-section through the spinal cord shows that gli3 (purple) is expressed in the neural tube dorsal to shh (red), which is in the floor plate (arrow) and notochord. (J) At 30 h, lateral view of dissected eye. gli3 is expressed weakly in the eye in cells surrounding the lens. (K–N) At 44 h. (K) At later stages, gli3 expression becomes restricted to the anterior telencephalon, dorsal diencephalon, hypothalamus, tectum, and dorsal hindbrain. Small clusters of cells also express gli3 in the ventral hindbrain. Mesendoderm ventral to the brain also expresses gli3 (arrow). (L) In the trunk, gli3 expression is restricted to the most dorsal regions of the spinal cord (arrowheads) and ventral endoderm (arrow). (M) Dorsal view of hindbrain showing two rows of ventral cells (see K for lateral view) on either side of the midline that express gli3. (N) gli3 is expressed throughout the fin bud with the exception of cells at the posterior margin (arrowhead). This expression overlaps with, but is much broader than, that of shh (right panel). (B, C, F–H, J–K) Side views, anterior to the left. (D, E, M, N) dorsal views. (I) Cross-section through trunk. (D–L) Yolks removed. (D, F, H) Eyes removed. Abbreviations: di; diencephalon, hb; hindbrain, hy; hypothalamus, tb; tail bud, t; telencephalon, tec; tectum. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.) |
|
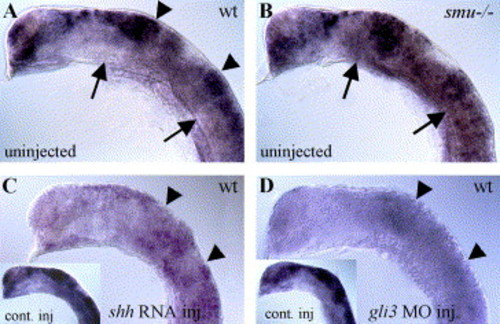
Hh signaling negatively regulates gli3 expression. (A) In 20-h wild-type embryos, gli3 is expressed regionally in the dorsal neural tube, with strong expression in the tectum and midbrain/hindbrain boundary (arrowheads). (B) gli3 expression is expanded ventrally in smu(smo) mutant embryos that lack Hh pathway activity (arrows). (C) gli3 expression is generally reduced in shh mRNA-injected wild-type embryos (compare to lacZ mRNA-injected embryo in inset). (D) gli3MO injection also led to severely reduced gli3 expression levels (compare to control MO-injected wild-type embryo in inset). All panels show gli3 expression in 20-h zebrafish embryos. Lateral views of head, anterior to the left, eyes and yolk removed. EXPRESSION / LABELING:
|
|
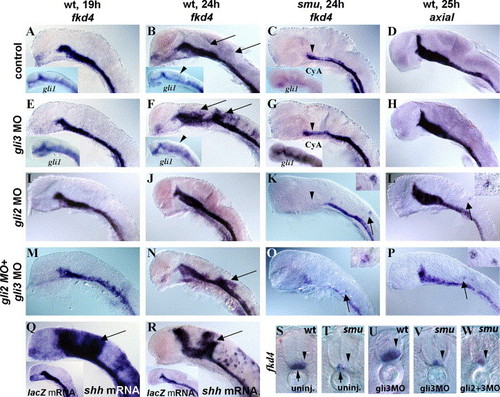
Temporally regulated Gli3 repression of the Hh target gene fkd4. (A, B, D) Control MO-injected wild-type embryos with normal expression of fkd4 (A, B) and axial (D) in the ventral brain, medial floor plate, and lateral floor plate. (E, F) gli3MO-injected wild-type embryos had normal expression of fkd4 and gli1 (inset) at 19 h (E) but had dorsally expanded fkd4 (F, U) and gli1 (F inset) expression at 24 h, revealing a temporally regulated repressor role for zebrafish Gli3. (C, G, T, V) smu(smo) mutant embryos had reduced fkd4 expression (C, T, arrowheads), and gli3MO injections into smu(smo) mutant embryos did not lead to dorsal expansion of fkd4 (G, V, arrowheads) indicating that active Hh signaling is necessary for this expansion. As in smu(smo) mutants (Karlstrom et al., 2003), cyclopamine treatment starting at shield stage led to an extreme reduction of gli1 expression at 20 h (C inset). gli3MO injection resulted in a dramatic increase in gli1 expression in cyclopamine-treated embryos (G inset). (D, H) No expansion of axial was detected in gli3MO-injected embryos (H), indicating target specificity for the Gli3 repressor. (I, J) gli2MO injection into wild-type embryos did not lead to expanded fkd4 expression either at 19 h (I) or at 24 h (J). (K, L) In contrast, a few cells ectopically expressed fkd4 in smu(smo) mutants (K) and axial in wild-type embryos (L) (see insets) after gli2MO injection, revealing a weak, Hh-independent, repressor function for Gli2. (M, N) Coinjection of gli3MOs and gli2MOs resulted in dorsal expansion of fkd4 in wild-type embryos at 24 h (N) but not 19 h (M), similar to single gli3MO injections. (O) Double gli2MO + gli3MO injections also led to ectopic fkd4 expression in a few dorsal neural cells in smu(smo) mutants at 24 h and expanded axial expression (P), similar to gli2MO injections alone. (Q, R) shh RNA-injected wild-type embryos had dorsally expanded fkd4 expression at both 19 h (Q) and 24 h (R) compared to lacZ mRNA-injected embryos (inset), showing that Shh signaling is capable of inducing fkd4 expression at both of these ages. (S) Cross-section shows fkd4 expression in the medial (arrow) and lateral (arrowhead) floor plate cells. (T) In smu(smo) mutants, fkd4 is expressed only in the medial floor plate (arrow) and is lost in lateral floor plate (arrowhead). (U) gli3MO injection into wild-type embryos led to dorsally expanded fkd4 expression. (V) gli3MO injection into smu(smo) mutant embryos did not rescue of fkd4 expression in lateral floor plate cells or expand fkd4 expression dorsally. (W) Similarly, injection of gli3MOs and gli2MOs into smu(smo) mutants did not rescue fkd4 expression in lateral floor plate or expand fkd4 expression dorsally. (A–R) Side views of zebrafish embryos, yolks and eyes removed, anterior to the left. (S–W) Cross-sections of the trunk region, anterior up. EXPRESSION / LABELING:
|
|
Positive regulation of nk2.2 and ptc1 expression by Gli3 and Gli1. (A) Wild-type nk2.2 expression in a control MO-injected zebrafish embryo. (B) In dtr(gli1) mutants nk2.2, expression is regionally absent (arrows). (C) smu(smo) mutants have no nk2.2 expression at 19 h. (D, E) gli3MO injections generally decreased nk2.2 expression in wild-type embryos (D) and further reduced nk2.2 expression in dtr(gli1) mutants (E, arrows). (F) Coinjections of gli3MO and gli1MO produced a smu(smo)-like phenotype with no nk2.2 expression. (G) ptc1 expression was not affected by control MO injections. (H) gli3MO injections had little or no effect on ptc1 expression. (I) gli1MO injections reduced expression of ptc1. (J) Coinjection of gli3MO and gli1MO reduced ptc1 expression almost completely. (A–F) lateral views of 19-h embryos, eyes and yolks removed, anterior to the left. (G–J) Dorsal views of 11-h embryos, anterior to the left, yolks removed. EXPRESSION / LABELING:
|
|
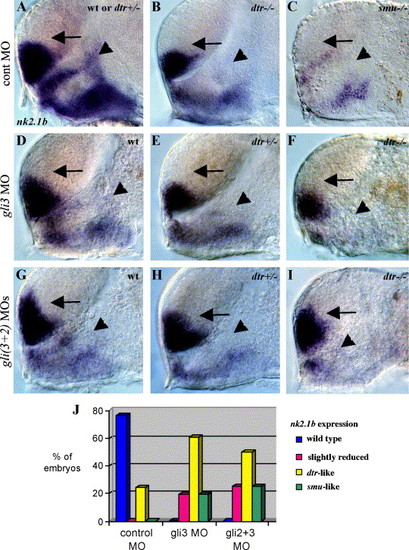
Regulation of nk2.1b expression by Gli3, Gli1, and Gli2. (A) nk2.1b expression in the forebrain is identical in wild-type and dtr(gli1)+/- heterozygous embryos. (B) dtr(gli1)-/- mutants have reduced nk2.1b expression in the dorsal diencephalon adjacent to the telencephalon (arrowhead). (C) smu(smo) mutant with greatly reduced nk2.1b expression in both the telencephalon and diencephalon. (D, E, F) gli3MO (10 ng) injected into a clutch of embryos from dtr(gli1)+/- heterozygous parents yielded 6 (26%) wild-type embryos which had a slight reduction of nk2.1b in the dorsal diencephalon (D, arrowhead), 10 (44%) heterozygous dtr(gli1)+/- embryos which had a dtr(gli1) mutant-like loss of nk2.1b expression in the dorsal diencephalon (E, arrowhead), and 7 (30%) dtr(gli1)-/- mutant embryos which had greatly reduced diencephalic nk2.1b expression (F, arrowhead) similar to phenotype seen in smu(smo) mutants. (G, H, I) Coinjection of gli3MOs and gli2MOs into a clutch of embryos from dtr(gli1)+/- heterozygous parents resulted in a slight reduction of diencephalic nk2.1b expression in 6 (25%) embryos (G, arrowhead), a dtr(gli1)-/- mutant-like reduction of nk2.1b expression in 12 (50%) embryos (dtr(gli1)+/- heterozygotes) (H, arrowhead), and greatly reduced diencephalic nk2.1b expression in 6 (25%) embryos (dtr(gli1)-/- mutants) (I, arrowhead). Double MO injections led to dorsal expansion of nk2.1b in the telencephalon (arrows in G, H, I), as shown previously for gli2MOs (Karlstrom et al., 2003). All panels show 30-h embryos, side views of the forebrain, eyes and yolk removed, anterior to the left. Genotype was determined by comparing defects in marker gene expression. (J) Graph showing percentages of injected embryos with different nk2.1b expression patterns. EXPRESSION / LABELING:
|
|
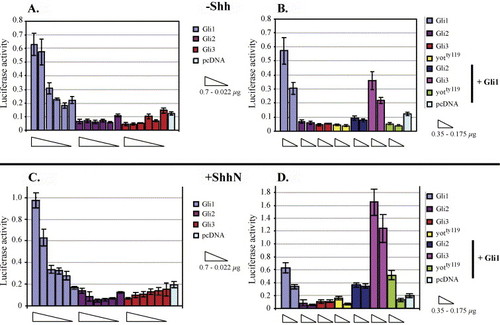
Transcriptional activity of zebrafish Gli1, Gli2, and Gli3 in C17 neuronal cells. (A–D) pcDNA constructs encoding zebrafish Gli1, Gli2, Gli3, and the yotty119 Gli2-dominant repressor (Gli2-DR) (Karlstrom et al., 2003) proteins were transfected into C17 cells along with a luciferase reporter gene under the transcriptional control of eight Gli1 binding sites (Sasaki et al., 1997). (A) In the absence of Shh, Gli1 (but not Gli2 or Gli3) can activate the reporter in a dose-dependent manner. (B) Cotransfection of multiple gli constructs in the absence of Shh shows that Gli2, truncated Gli2 proteins encoded by zebrafish you-too mutants, and Gli3 can all repress Gli1-mediated transcriptional activity. (C, D) The same experiments were performed in a cell line stably expressing Shh (Feng et al., 2004). (C) In the presence of Shh, Gli1 activator function is largely unaltered, and Gli2 and Gli3 do not activate the reporter construct. (D) In the presence of Shh, Gli3 expression enhances rather than represses Gli1-mediated reporter gene expression. EXPRESSION / LABELING:
|
|
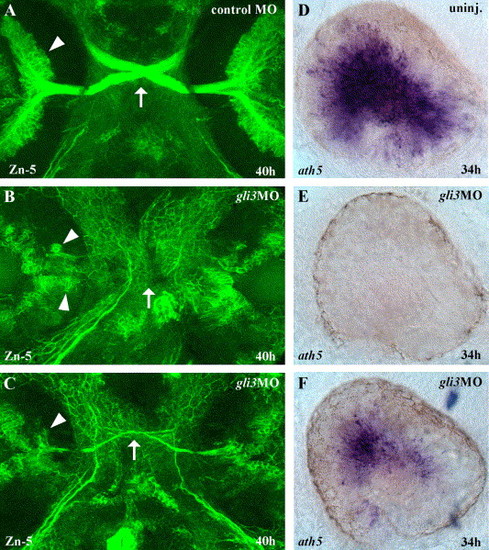
Gli3 is required for RGC differentiation. (A) At 40 h of development, large numbers of Zn-5 labeled retinal ganglion cells (RGCs) (arrowheads) in the eyes have differentiated and extended axons across the midline to form the optic nerve and chiasm (arrow). (B) In severely affected gli3MO injected embryos, very few RGCs differentiated, and those that were present were distributed aberrantly in the eye (arrowheads). (C) In less severely affected gli3MO injected embryos, reduced numbers of RGCs differentiated in the correct location and extended axons across the midline to form a thin optic nerve and chiasm (arrow). (D) At 34 h, the zebrafish atonal homolog ath5 is expressed throughout the central region of the differentiating retina. (E) In severely affected gli3MO-injected embryos, ath5 expression was completely eliminated in the eye. (F) In less severely affected gli3MO-injected embryos, ath5 expression was strongly reduced. (A–C) Fluorescent labeling of RGCs at 40 h using the Zn-5 antibody, ventral views of head, anterior up. (D–F) ath5 in situ labeling of the eyes at 34 h, lateral views of the left eye, dorsal up, anterior to the left. |

Unillustrated author statements EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 277(2), Tyurina, O.V., Guner, B., Popova, E., Feng, J., Schier, A.F., Kohtz, J.D., and Karlstrom, R.O., Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling, 537-556, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.