Fig. 5.
- ID
- ZDB-FIG-230211-78
- Publication
- Hajiabadi et al., 2022 - Deep-learning microscopy image reconstruction with quality control reveals second-scale rearrangements in RNA polymerase II clusters
- Other Figures
- All Figure Page
- Back to All Figure Page
|
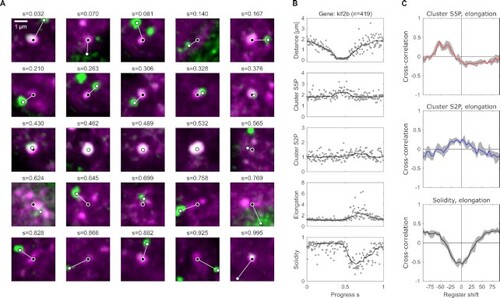
Pseudo-time analysis of data from fixed embryos relates transient engagement and activation of a gene to the phosphorylation and shape changes observed in live embryos. (A) Example images of Pol II Ser5P (magenta signal) clusters sorted by a pseudo-time progress coordinate ( |