Fig. 1.
- ID
- ZDB-FIG-230211-75
- Publication
- Hajiabadi et al., 2022 - Deep-learning microscopy image reconstruction with quality control reveals second-scale rearrangements in RNA polymerase II clusters
- Other Figures
- All Figure Page
- Back to All Figure Page
|
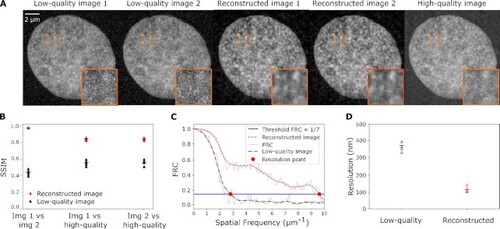
Metrics for the reliability and effective resolution in n2v-reconstructed images. (A) Representative micrographs of the DNA distribution in a nucleus in a fixed zebrafish embryo, recorded with a stimulated emission depletion (STED) super-resolution microscope. The same image plane was recorded twice at low quality, once at high quality, and two n2v-reconstructed images were prepared from the low-quality images. (B) SSIM values for pair-wise comparison (image 1 vs. image 2) and comparison against the high-quality image (image 1 vs. high-quality and image 2 vs. high-quality) for the low-quality images and the reconstructed images. (C) FRC curves calculated based on a low-quality image pair and the corresponding reconstructed image pair. (D) FRC-based effective resolution for four pairs of low-quality images and the corresponding pairs of reconstructed images. |