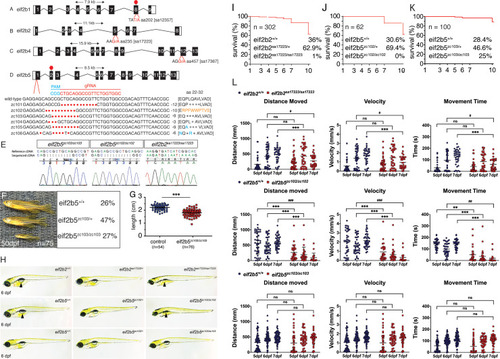
(A-D) Depiction of zebrafish eif2b subunits exon structure and the location and nucleotide change for each mutant. (A) eif2b1 harbors a T/A transversion resulting in an early stop in exon 8. (B) eif2b2 has a G/A transition in exon 5, mutating an essential splice site. (C) eif2b4 has a G/A transition in exon 12 mutating an essential splice site. (D) eif2b5 exon one was targeted for mutagenesis using a gRNA (red). Six distinct alleles were recovered (described in text). (E) Chromatograms of cDNA confirm presence of predicted mutations for eif2b5zc103/zc103, eif2b5zc102/zc102, and eif2b2sa17223/sa17223. (F) eif2b5zc103/zc103 mutants survive until adulthood in Mendelian ratios, but show grow defects compared to their heterozygous and wild-type siblings. (G) Adult eif2b5zc103/zc103 lengths are significantly shorter compared to their wild-type and heterozygous siblings. (H) Bright-field (BF) images of 6 dpf eif2b5zc103/zc103, eif2b5zc102/zc102, and eif2b2sa17223/sa17223 larva. eif2b2sa17223/sa17223 and eif2b5zc102/zc102 have no swim bladder (arrowhead) and a small head. (I) Kaplan-Meyer survival curves from an eif2b2sa17223/+ heterozygous in-cross shows 1% (n = 3) homozygote survival at 10 dpf (total n = 302); however no homozygotes live past 2 weeks of age. (J) Kaplan-Meyer survival curves from an eif2b5zc102/+ heterozygous in-cross shows that all homozygotes were dead by 10 dpf (total n = 62). (K) Kaplan-Meyer survival curves from an eif2b5zc103/+ heterozygous in-cross show no mortality of homozygotes. (L) Motor swimming analysis shows impaired swimming behavior in mutants. Distance moved, time spent moving, and velocity, for wild-type controls, and eif2b5zc103/zc103, eif2b5zc102/zc102, and eif2b2sa17223/sa17223 mutants, at 5, 6, and 7 dpf. Mean shown with 95% confidence intervals.
|

