- Title
-
A targeted CRISPR-Cas9 mediated F0 screen identifies genes involved in establishment of the enteric nervous system
- Authors
- Moreno-Campos, R., Singleton, E.W., Uribe, R.A.
- Source
- Full text @ PLoS One
|
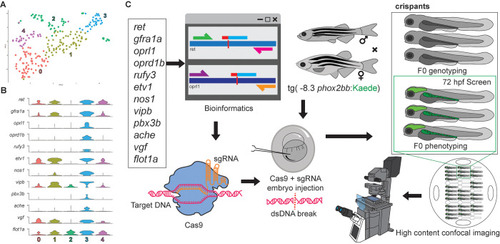
construction of an F0 CRISPR screen for ENS development. (A) tSNE plot shows five distinct sub-clusters after the subset analysis and re-clustering of Clusters 5 and 12 from the [ |
|
Candidate genes targeted in an F0 CRISPR screen display ENS phenotypic alterations. (A) At 48 hpf, eight crispant embryos from pools of around thirty embryos were used to validate CRISPR activity via T7E1, for each gene targeted. If the majority had indels, then subsets of the pool were grown at 3 to 4 dpf to phenotype their ENS. The phenotyping process combined crispants of different genes by using an agarose cast that enabled high-content semi-automated confocal imaging. An additional fraction of the crispants were analyzed at 4–6 dpf for additional HCR validation or for late phenotypic alterations. (B) Representative images of different T7E1 assays demonstrate indels of different embryos in the specific gene-targeted regions. |
|
Expression pattern of CRISPR screen selected genes along the ENS during development. (A, C, E, G) Confocal images show HCR-assayed expression for |
|
Temporal chemical inhibition of opioid receptors induces ENS developmental defects in zebrafish larvae. (A) Tg(-8.3 |
|
ENS neurochemical coding is altered in larval crispants for opioid receptor-encoding genes (A, C, E) Confocal images show whole ENS after immunohistochemistry in 6 dpf control, and |