- Title
-
miR-34a is a tumor suppressor in zebrafish and its expression levels impact metabolism, hematopoiesis and DNA damage
- Authors
- Prykhozhij, S.V., Ban, K., Brown, Z.L., Kobar, K., Wajnberg, G., Fuller, C., Chacko, S., Lacroix, J., Crapoulet, N., Midgen, C., Shlien, A., Malkin, D., Berman, J.N.
- Source
- Full text @ PLoS Genet.
|
Conservation, genomic synteny and expression timing of microRNA-34 family members in zebrafish. |
|
p53 induces all miR-34 genes in zebrafish but with different kinetics. ( |
|
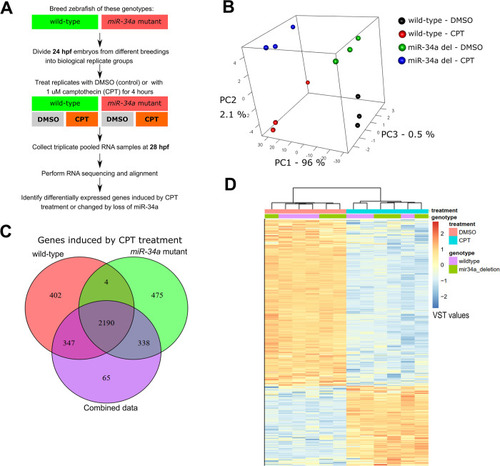
p53 activation by camptothecin treatment induces massive gene expression effects with a small contribution from |
|
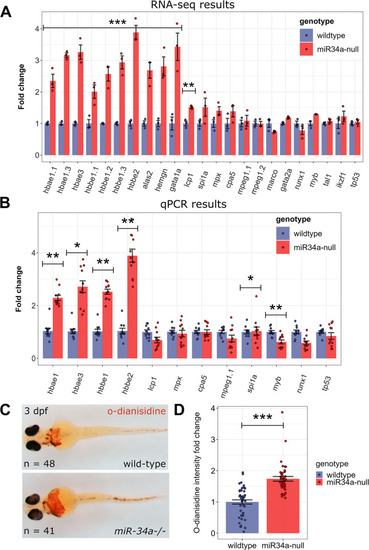
Expression changes due to loss of |
|
Tumor development due to |
|
Expression profiling by RNA-seq of wild-type vs |
|
Analysis of blood cell type markers in 3 dpf wild-type and |
|
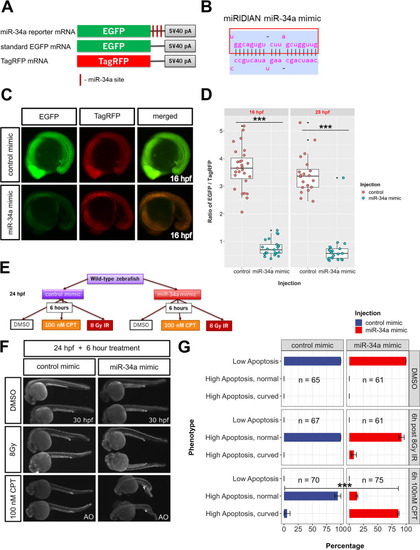
Transient overexpression of miR-34a sensitizes zebrafish embryos to camptothecin treatment. |