- Title
-
Identification of Dhx15 as a Major Regulator of Liver Development, Regeneration, and Tumor Growth in Zebrafish and Mice
- Authors
- Portolés, I., Ribera, J., Fernandez-Galán, E., Lecue, E., Casals, G., Melgar-Lesmes, P., Fernández-Varo, G., Boix, L., Sanduzzi, M., Aishwarya, V., Reig, M., Jiménez, W., Morales-Ruiz, M.
- Source
- Full text @ Int. J. Mol. Sci.
|
|
|
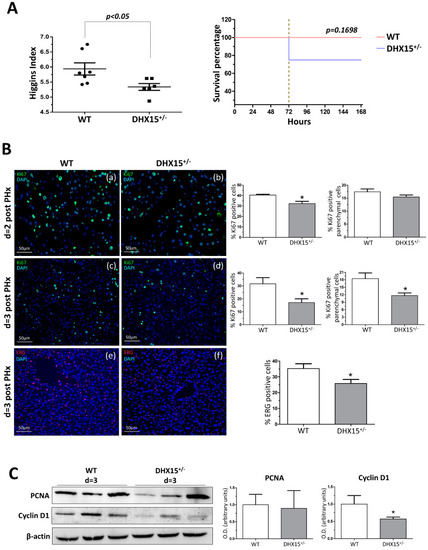
Intrahepatic liver vasculature was altered in |
|
|
|
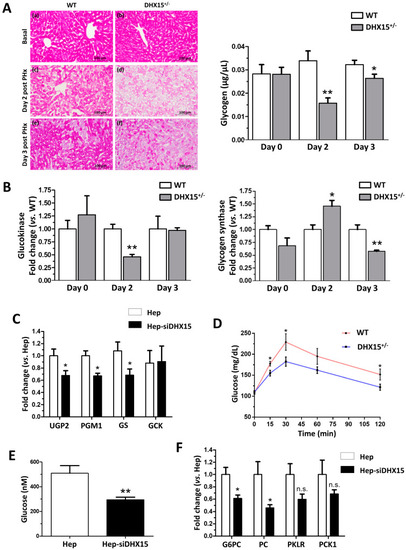
Impaired glucose metabolism in |
|
Tumor growth and metastases in |
|
Tumor growth in wild-type mice following Hepa1-6 tumor induction after |