- Title
-
Identification of kinase modulators as host-directed therapeutics against intracellular methicillin-resistant Staphylococcus aureus
- Authors
- van den Biggelaar, R.H.G.A., Walburg, K.V., van den Eeden, S.J.F., van Doorn, C.L.R., Meiler, E., de Ries, A.S., Meijer, A.H., Ottenhoff, T.H.M., Saris, A.
- Source
- Full text @ Front Cell Infect Microbiol
|
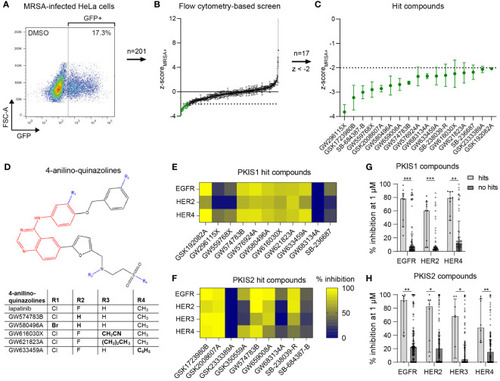
Identification of PKIS compounds inhibiting intracellular growth of MRSA. |
|
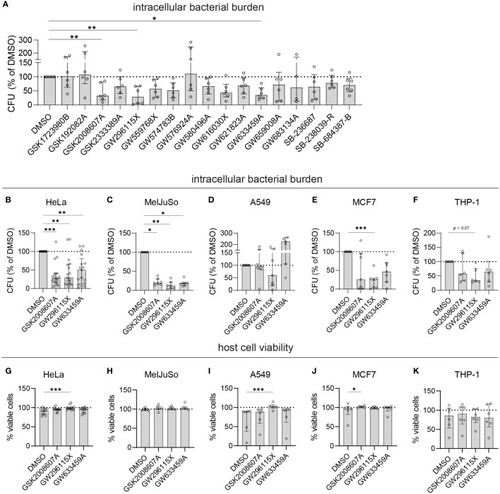
Validation of selected PKIS hit compounds. |
|
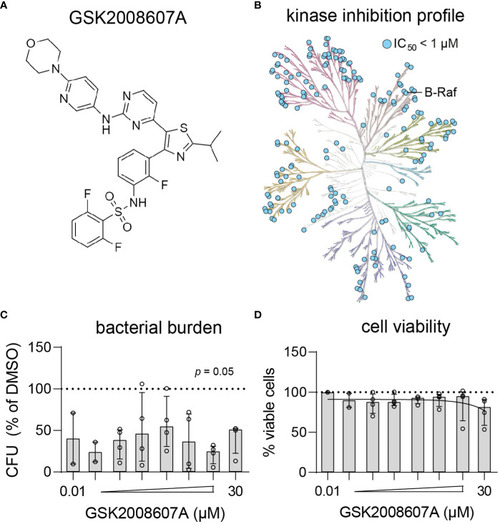
Kinase inhibition profile and potency of GSK2008607A. |
|
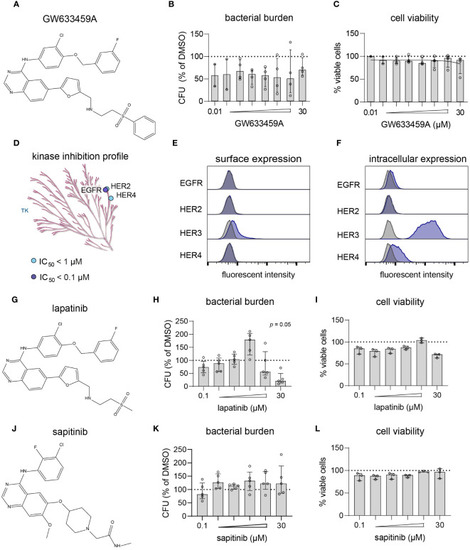
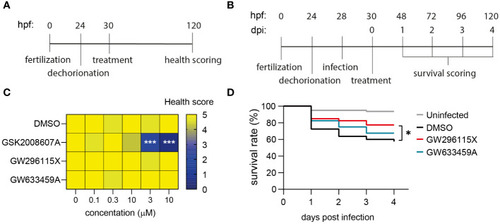
GW633459A is an inhibitor of the HER kinase family and reduces intracellular MRSA burden, but probably through a mechanism unrelated to these host kinase targets. |
|
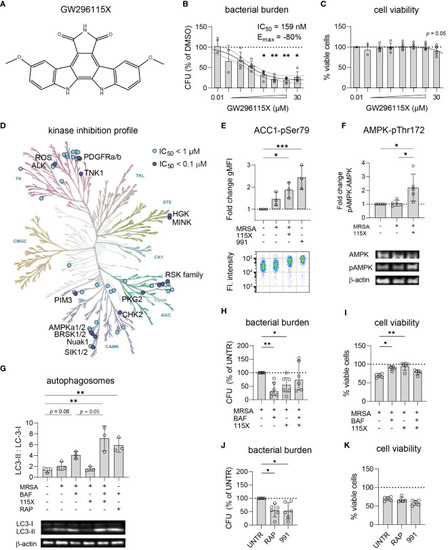
GW296115X activates AMPK resulting in autophagosome formation and elimination of intracellular MRSA. |
|
Safety and efficacy of PKIS hit compounds in |
|
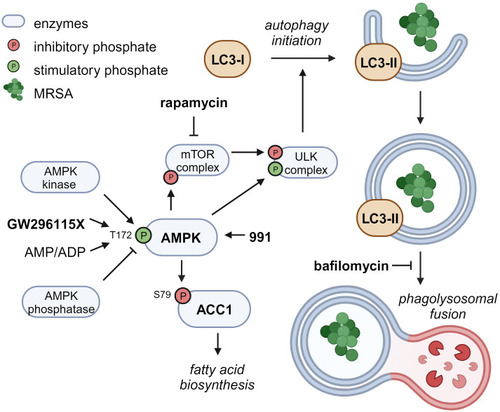
Proposed mechanism of action of compound GW296115X. Compound GW296115X either stimulates AMPK T172 phosphorylation or inhibits its dephosphorylation, resulting in activation of AMPK. Active AMPK phosphorylates ACC1, resulting in decreased fatty acid biosynthesis. In addition, active AMPK phosphorylates the mTOR complex and the ULK complex both directly, thereby inactivating the mTOR complex and activating the ULK complex, which facilitates the initiation of autophagy. The AMPK-activating 991 and mTOR-inhibitor rapamycin show similar effects on ACC1 phosphorylation and autophagy initiation, respectively. Bafilomycin blocks autophagosome degradation, thereby resulting in increased LC3-II levels, especially in combination with drugs that stimulate autophagy initiation including GW296115X or rapamycin. Created with |