- Title
-
Forward genetic screen using a gene-breaking trap approach identifies a novel role of grin2bb-associated RNA transcript (grin2bbART) in zebrafish heart function
- Authors
- Angom, R.S., Joshi, A., Patowary, A., Sivadas, A., Ramasamy, S., K V, S., Kaushik, K., Sabharwal, A., Lalwani, M.K., K, S., Singh, N., Scaria, V., Sivasubbu, S.
- Source
- Full text @ Front Cell Dev Biol
|
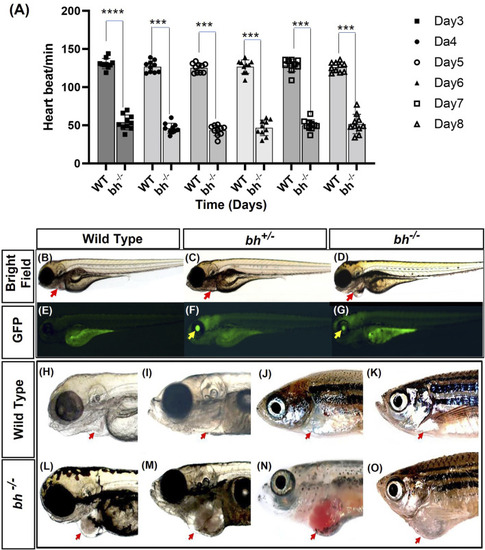
Heartbeat rate analysis and phenotypic characterization of |
|
The electrocardiogram profile of |
|
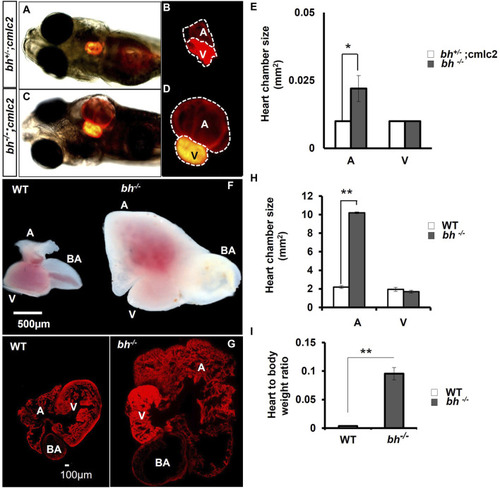
Anatomical study of |
|
|
|
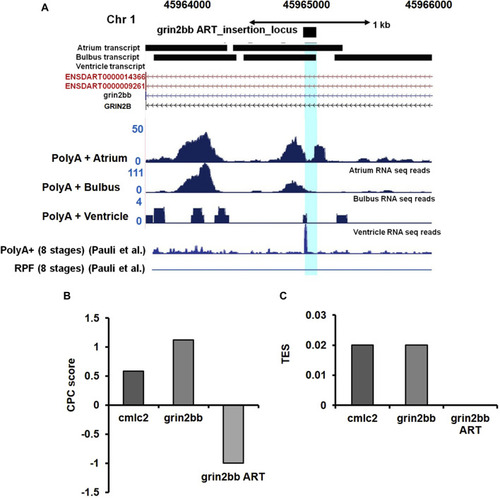
Ultra-deep RNA sequencing of WT heart suggests the presence of unannotated transcript in |
|
|
|
Whole mount |
|
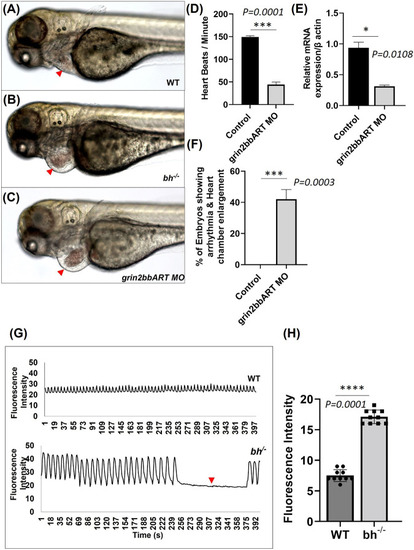
Big heart mutants displayed calcium mishandling and morpholino based knockdown of |
|
Transcriptome analysis of bigheart mutant. |