- Title
-
Defining morphologically and genetically distinct GAGBergic/cholinergic amacrine cell subtypes in the vertebrate retina
- Authors
- Li, Y., Yu, S., Jia, X., Qiu, X., He, J.
- Source
- Full text @ PLoS Biol.
|
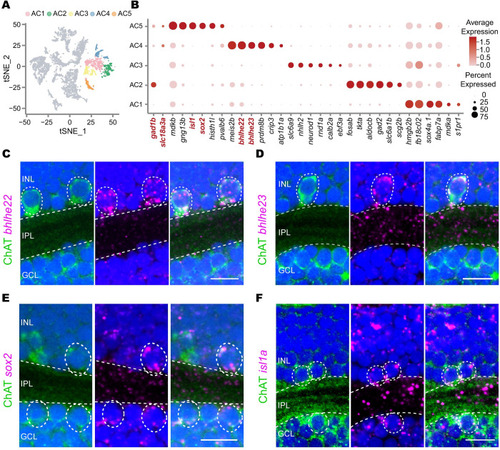
Single-cell RNA-seq identifies 2 GABAergic/cholinergic AC types. ( |
|
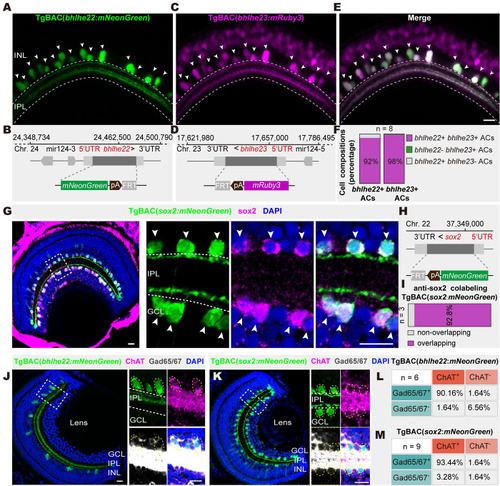
Genetically marking 2 GABAergic/cholinergic AC types. ( |
|
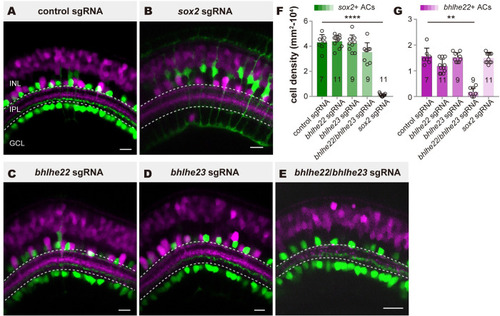
The generation of 2 AC types requires distinct sets of TFs. ( |
|
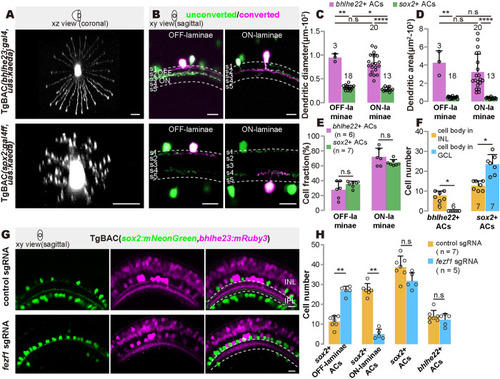
Morphological characteristics of 2 GABAergic/cholinergic AC types. ( |
|
Direction selectivity of 2 GABAergic/cholinergic AC types. ( |
|
( |