- Title
-
Zebrafish null mutants of Sept6 and Sept15 are viable but susceptible to Shigella infection
- Authors
- Torraca, V., Bielecka, M.K., Gomes, M.C., Brokatzky, D., Busch-Nentwich, E.M., Mostowy, S.
- Source
- Full text @ Cytoskeleton
|
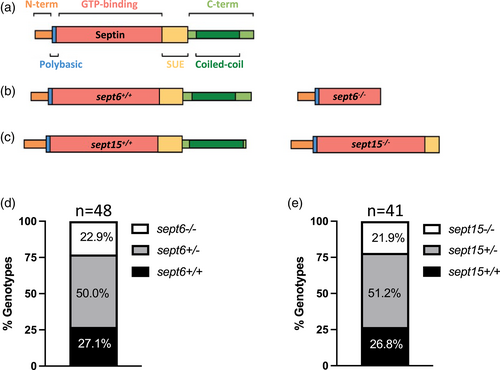
Generation of sept6 and sept15 mutants. (a) Schematic representation of the general septin protein structure and consequence of gene mutations at the level of the protein structure. (b,c) Specific representation of Sept6 (b) and Sept15 (c) protein structure and consequence of gene mutations at protein levels in Sept6 and Sept15 mutants. A sept6 null allele was generated by CRISPR/Cas9 gene editing. A sept15 null allele was identified from an ENU mutagenesis screen. (d,e) Genotyping of the adult offspring of sept6 (d) and sept15 (e) heterozygote carriers indicated no inheritance disequilibrium and homozygote mutants reached adulthood at the same rate as co-housed wild-type fish. The proportions of the different genotypes did not significantly differ from the attended Mendelian proportions by chi-squared test. Sample size: a total of 48 (sept6) and 41 (sept15) adults (~6 months old) were genotyped PHENOTYPE:
|
|
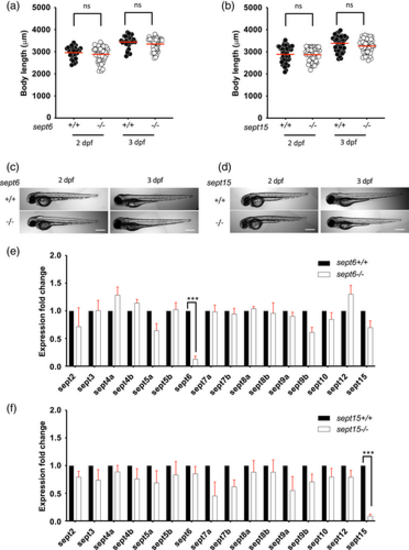
Characterisation of septin mutants. (a–d) sept6 (a,c) and sept15 (b,d) mutants do not have aberrant development. Comparison of the larvae length for sept6 (a) and sept15 (b) mutants to wild-type larvae, at 2 and 3 dpf. The body lengths of mutants and wild-types did not differ significantly. Representative images of sept6 (c) and sept15 (d) mutant and wild-type larvae at 2 and 3 dpf. Scale bar: 500 μm. Results are cumulative of three independent experiments. Sample size: in total, 30 (wild-type) and 48 (mutant) fish larvae were analysed for sept6 and 48 (wild-type) and 48 (mutant) fish larvae were analysed for sept15. Statistics: Kruskal–Wallis test; ns p > 0.05. (e,f) Both sept6 (e) and sept15 (f) homozygote mutations lead to nonsense-mediated mRNA decay of the mutated transcript but do not significantly impact the expression level of other septins. Expression analysis was performed by qRT-PCR, using primers able to amplify both mutant and wild-type transcripts. Data are represented as expression fold changes against the expression level of the wild-type group. Sample size: 15–20 larvae at 3 dpf were pooled together to extract mRNA from four independent replicates. Statistics: differences in gene expression levels were determined by multiple paired t tests, with Benjamini, Krieger, and Yekutieli to correct for false discovery (false discovery rate assumed 1%). ***p < 0.001 EXPRESSION / LABELING:
PHENOTYPE:
|
|
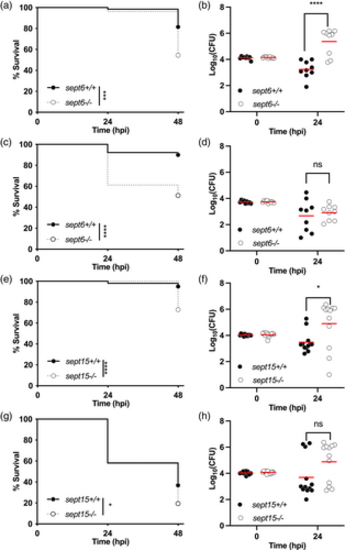
Zebrafish null mutants of Sept6 and Sept15 are more susceptible to Shigella infection. (a–d) Survival curves (a,c) and Log10-transformed CFU counts (b,d) of 3 dpf sept6+/+ and sept6−/− larvae (wild-type in black; mutant in white) injected in the hindbrain ventricle (HBV) with S. flexneri GFP+ at a dose of 10,000 CFUs (a,b) or a dose of 5,000 CFUs (c,d). Next, larvae were incubated at either 28.5°C (a,b) or 32.5°C (c,d), for up to 48 h. Experiments are cumulative of three biological replicates. Sample size: For survival analysis, a total of 65 (wild-type) and 79 (mutant) and 90 (wild-type) and 80 (mutant) larvae was analysed at 28.5°C and 32.5°C, respectively. For CFU analysis, a total of 9 larvae was analysed per experimental group. Only living larvae were used for CFU enumeration. Statistics: Log-rank (Mantel-Cox) test (a,c); unpaired t test on Log10-transformed values (b,d). ****p < 0.0001; ns p > 0.05, respectively. (e,h) Survival curves (e,g) and Log10-transformed CFU counts (f,h) of 3 dpf sept15+/+ and sept15−/− larvae (wild-type in black; mutant in white) injected in the HBV with S. flexneri GFP+ at a dose of 10,000 CFUs. Next, larvae were incubated at either 28.5°C (e,f) or 32.5°C (g,h) for up to 48 h. Experiments are cumulative of four biological replicates. Sample size: For survival analysis, a total of 99 (wild-type) and 104 (mutant) and 103 (wild-type) and 113 (mutant) larvae was analysed at 28.5°C and 32.5°C, respectively. For CFU analysis, a total of 12 larvae was analysed per experimental group. Only living larvae were used for CFU enumeration. Statistics: Log-rank (Mantel–Cox) test (e, g); unpaired t test on Log10-transformed values (f,h). *p < 0.05; ****p < 0.0001; ns p > 0.05, respectively PHENOTYPE:
|