- Title
-
Sirt1 promotes tissue regeneration in zebrafish through regulating the mitochondrial unfolded protein response
- Authors
- Lin, Y.F., Sam, J., Evans, T.
- Source
- Full text @ iScience
|
Figure 1. The UPRmt is activated in adult and larval injured fins (A and B) Representative in situ hybridization images (left panels) show that lonp1 (a UPRmt gene) is up-regulated during fin regeneration in adult (A, partial fin view, posterior toward top) and larval (B, partial body view, posterior toward the right) fish at 2 dpa or 4 hpa, respectively. qRT-PCR assays (right panels) show that the expression of all four UPRmt signature genes are enhanced in adult and larval injured fin tissue. (C-i) Diagram indicates the hspd1 genomic fragment (2.6 kb upstream of the transcription start (+1) to 3 kb downstream until the translation start) employed for driving the expression of the destabilized GFP. (C-ii and C-iii) Adult fin of the UPRmt reporter fish, Tg(hspd1:d2gfp), exhibits induced fluorescence at the injury zone during regeneration. Higher magnification image (iii; white rectangle in ii) indicates that the fluorescent signal marks the blastema of the injured fin. (C-iv to C-vi) A time series of images shows the dynamics of the UPRmt reporter activation in larval fin upon injury. Note fluorescent migrating mesenchymal-like cells. (D) Representative images of hspd1 in situ hybridization assays show that hspd1 expression is diminished in SU5402 treated injured larvae at 4 hpa. (Error bars indicate standard deviation. Student’s t test was performed to determine statistical significance. ∗p < 0.05). Scale bars are as indicated. EXPRESSION / LABELING:
PHENOTYPE:
|
|
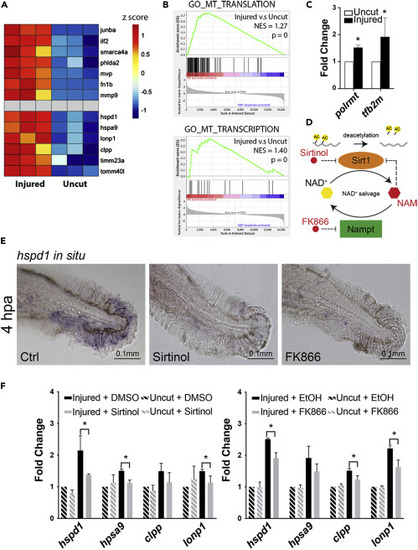
Figure 2. Chemical inhibition of Sirt1 impairs UPRmt activation in injured larval fins (A) Heatmap of relative gene expression levels derived from the transcriptional profiling data demonstrates that previously studied regeneration genes (top half) as well as the UPRmt genes (bottom half) are all up-regulated in the injured fins during regeneration. (B) GSEA indicates that mitochondrial translation and transcription are among the cellular processes that are most enhanced during fin regeneration. (C) qRT-PCR assays show that the expression of two mitochondrial transcriptional machinery components, polrmt and tf2bm, are enhanced in zebrafish adult fins during regeneration. (D) The schematic depicts the mechanism of Sirt1 inhibition by various small molecule inhibitors used in subsequent experiments. (E) Representative images of hspd1 in situ hybridization. (F) qRT-PCR assays show that Sirt1 inhibition via Sirtinol treatment or Nampt inhibition via FK866 treatment suppresses the UPRmt gene expression in the injured larval fin. (Error bars indicate standard deviation. Student’s t test was performed to determine statistical significance. ∗p < 0.05.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
Figure 3. Sirt1 inhibition causes a fin regeneration defect in larval fish (A) Representative images of larval fins show the outcomes of the regenerative fin growth in the control and the Sirtinol + NAM treatment conditions at 3 dpa. (The white dashed line in the left panel indicates the resecting line.) The quantification in the graph on the right shows that Sirt1 inhibition by Sirtinol alone or combination of Sirtinol or FK866 with NAM significantly impairs regenerative fin growth in larval fish. (B) The quantification shows that Sirt1 inhibition by a high concentration of NAM significantly impairs regenerative fin growth in larval fish. (C) The schematic shows the alignment of genomic DNA sequence of the wild type and a sirt1 mutation containing a 28 bp insertion in exon 1 generated by CRISPR/Cas9, which results in a premature stop codon (indicated by an asterisk). The official allele name for this mutation is sirt1wcm18/wcm18. The agarose gel image shows that PCR of genomic DNA generates a larger band from a homozygous sirt1 mutant fish compared to wildtype (WT). (D) Representative images (left) and the quantification result (right) show that sirt1 mutant fish fail to regenerate fins when incubated in NAM. Each dot represents fin regeneration measurement from one fish. Error bars indicate standard deviation. Student’s t test was performed to determine statistical significance. ∗p < 0.05.) PHENOTYPE:
|
|
Figure 4. Sirt1 inhibition impairs adult fin regeneration via reduction of the UPRmt (A) Representative images (left) and quantification (right) show that inhibiting Sirt1 by adding NAM to fish water impairs fin regeneration in adult zebrafish. (B) Representative in situ hybridization images show that NAM treatment reduces hspd1 expression levels in the blastema of an injured adult fin. (C) The representative images (left) and quantification (right) show that incubation of injured hspd1 mutant fish at a semi-permissive temperature in conjunction with a low concentration of NAM results in significant inhibition of fin regeneration. (D) Quantification of regeneration growth shows that sirt1 mutant larval fish exhibit a fin regeneration defect when treated with doxycycline. Each dot represents a fin regeneration measurement from one fish. Error bars indicate standard deviation. Student’s t test was performed to determine statistical significance. ∗p < 0.05.). PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|