- Title
-
Analysis of gene network bifurcation during optic cup morphogenesis in zebrafish
- Authors
- Buono, L., Corbacho, J., Naranjo, S., Almuedo-Castillo, M., Moreno-Marmol, T., de la Cerda, B., Sanbria-Reinoso, E., Polvillo, R., Díaz-Corrales, F.J., Bogdanovic, O., Bovolenta, P., Martínez-Morales, J.R.
- Source
- Full text @ Nat. Commun.
|
EXPRESSION / LABELING:
|
|
|
|
|
|
|
|
|
|
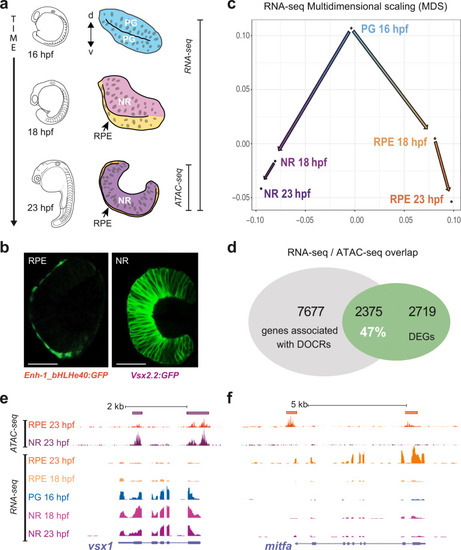
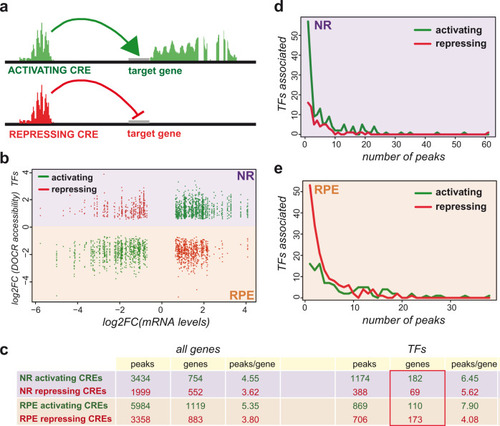
Representative TF motifs enriched in activating (left column) and repressing (right column) CREs in both the NR and RPE domains. Analysis of GO terms enrichment for genes associated with each set of elements is indicated. |
|
EXPRESSION / LABELING:
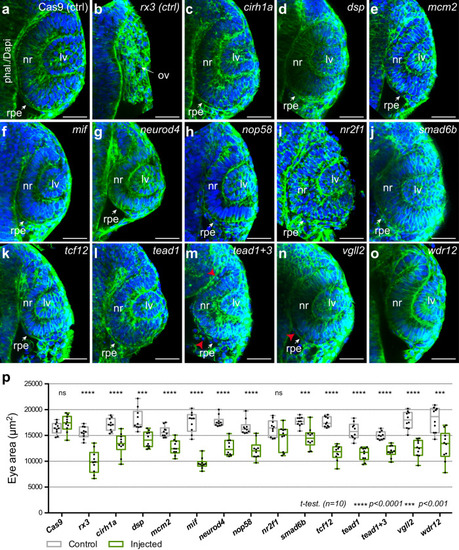
PHENOTYPE:
|
|
PHENOTYPE:
|
|
|