- Title
-
Zebrafish model of KRAS-initiated pancreatic cancer
- Authors
- Park, J.T., Leach, S.D.
- Source
- Full text @ Animal Cells Syst (Seoul)
|
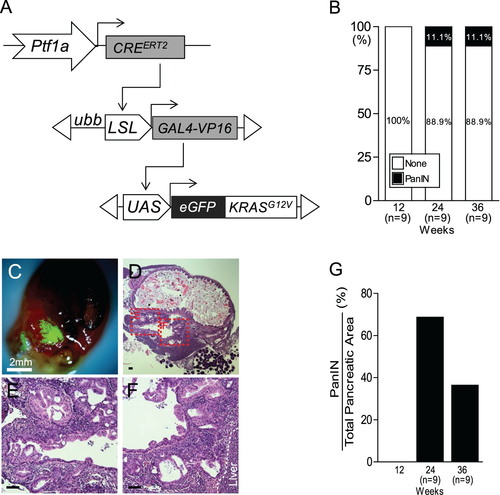
Identification of tge PanIN region in Tg (ptf1a:CREERT2; LSL-GAL4; UAS-KRASG12V) fish. (A) Schematic of the ptf1a CRE-driver line,Tg (ptf1a:CREERT2), the CRE-responder line Tg (LSL-GAL4), and the GAL4-responder line Tg (UAS-KRASG12V). (B) Quantification of PanIN induction frequency in Tg (ptf1a:CREERT2; LSL-GAL4; UAS-KRASG12V) fish. (C) Dissected abdominal viscera with an eGFP-positive tumor from KRASG12V. Scale bars: 2 mm. (D–F) The histological profiles of tumors bear striking resemblance to human PanIN. Boxed areas indicate regions depicted at higher magnification in adjacent images. Scale bars: 50 μm. (G) Quantification of PanIN region vs. total pancreatic area in Tg (ptf1a:CREERT2; LSL-GAL4; UAS-KRASG12V) fish. PHENOTYPE:
|
|
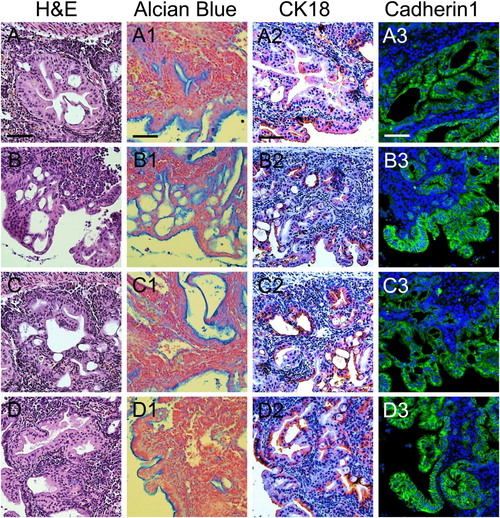
Histological and immunohistochemical profiles in PanIN regions. (A–D) Several grades of differentiation ranging from PanIN-1 to PanIN-3 in PanIN regions, as indicated by hematoxylin and eosin (H&E) staining. (A1–D1) Alcian blue staining. (A2–D2) CK-18 staining. (A3–D3) Cadherin1 staining PHENOTYPE:
|
|
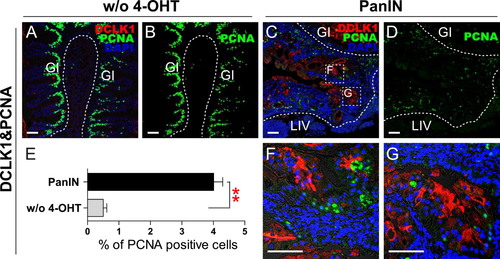
DCLK1 and PCNA staining in PanIN regions. (A and B) DCLK1 and PCNA staining in groups without 4-OHT treatment (w/o 4-OHT) as controls. Scale bars: 50 μm. (C and D) DCLK1 and PCNA staining in PanIN regions. Boxed areas indicate regions depicted at higher magnification in adjacent images. Scale bars: 50 μm. (E) Quantification of PCNA positive cells in groups without 4-OHT treatment, and in PanIN regions. (**P < .01, one-way ANOVA). (F and G) Magnified region of boxed areas in Figure 3(C). Scale bars: 50 μm. |
|
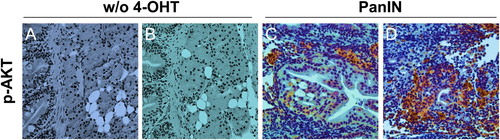
Characterization of the KRAS downstream signaling pathway in PanIN regions. (A and B) Phospho-AKT staining in groups without 4-OHT treatment (w/o 4-OHT) as controls. Scale bars: 50 μm. (C and D) Phospho-AKT staining in PanIN regions. Scale bars: 50 μm. |