- Title
-
Zebrafish snai2 mutants fail to phenocopy morphant phenotypes
- Authors
- Bickers, C., Española, S.D., Grainger, S., Pouget, C., Traver, D.
- Source
- Full text @ PLoS One
|
Snai2 endogenous expression in wild-type embryos. Expression of snai2 was analyzed via whole mount in situ hybridization at 14 hpf (A), 18 hpf (B), and 24 hpf (C). Embryos were cryosectioned post-in situ at 18 hpf (D) and 24 hpf (F). Simplified schematics are provided (E and G). Double fluorescent in situ for snai2 with endothelial markers fli1a at 14 hpf (H) and etsrp at 26 hpf (I) was performed. Insets show a close-up view of the PLM. QPCR was used to compare snai2 enrichment within double positive HSPCs sorted from Tg(CD41:GFP/kdrl:mCherry) on 2 dpf to the rest of the embryo. Markers cmyb and kdrl, which should be enriched in this population, are displayed alongside for comparison. N: notochord; NT: neural tube; S: somite. EXPRESSION / LABELING:
|
|
Snai2 morphants display a strong defect in HSC specification. Expression of the HSC specification marker, runx1, was analyzed by in situ hybridization at ~26 hpf in embryos injected with SB MO, ATG MO, and their siblings (A). The effect of snai2 mRNA injection was also analyzed both alone and when coinjected with SB MO. Black arrowheads point to the middle of the aortic runx1 expression. Numbers in the lower right hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. Tg(gata2b:Gal4/UAS:LA-GFP/kdrl:mCherry) morphants and their siblings were imaged by confocal microscopy at 48 hpf, and Imaris imaging software was used to remove GFP signal outside of the vasculature (B). Pink coloration is indicative of double positive cells as filtered by the surfaces feature of Imaris. White arrowheads indicate separate putative HSPCs. Quantification for each fish was graphed and statistically analyzed by non-parametric t-test on Prism (C). Error bars are SEM. |
|
Snai2 SB morphants display depletion of sclerotome markers. pax9, a marker of the sclerotome, was analyzed by WISH at 18 hpf (A). Black brackets highlight the difference in staining between morphants and siblings. qPCR on somitic, GFP+ cells sorted from morphant and control Tg(actc1b:GFP) embryos at ~17 hpf showed the sclerotome marker twist1b was decreased, while the pan-somitic marker remained normal in morphants (B). Wish for foxc1b showed clear diagonal expression in the anterior portion of the somites of uninjected embryos, while morphants lacked this distinctive stripe pattern (C). Error bars are calculated from technical replicates. Double fluorescent in situ hybridization for foxc1b and myoD (myogenic marker) followed by razor cutting for confocal analysis showed that while the muscle marker is consistent in both morphants and uninjected embryos, there is a notable decrease of positive staining for foxc1b, especially within the dorsal portion of the somites. A small schematic is provided to show greater detail of how embryos are oriented in Fig D. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Snai2 SB morphants have defective Notch signaling. Aortic Notch activity was assessed by confocal microscopy of the Notch reporter Tg(TP1:GFP) (A). Median fluorescence intensity was calculated by the surfaces feature of Imaris and was graphed and statistically analyzed by a non-parametric t-test on Prism (B). Error bars are SEM. Using a combination of Tg(kdrl:miniGal4) and Tg(UAS:NICD-myc), we saw that ectopically activating Notch signaling within the endothelium was sufficient to rescue expression of the HSC marker runx1 in morphant embryos (C). Black arrowheads point to the middle of the aortic runx1 expression. Analysis by WISH displays that expression of the Notch ligands dlc and dld is decreased in morphants, especially within the more anterior somites (D). Black brackets are provided to highlight the differences in staining. This decrease was further confirmed by qPCR in somitic, GFP+ cells sorted from morphant Tg(actc1b:GFP) embryos as compared to uninjected siblings (E). Snai2 relative expression is included for comparison, since the misspliced transcript is consistently elevated in SB MO injected embryos. Error bars are calculated from technical replicates. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Snai2112Δ and snai2sa24539 mutants have no embryonic defects in HSC or sclerotome formation. WISH analysis was performed on embryos derived from heterozygote in-crosses including probing for the hematopoietic markers runx1 (A) and cmyb (B) and the sclerotome marker foxc1b (C). Wild-type, heterozygotes, and mutants are all included for the Snai2112Δ allele since genotyping without sequencing was possible. For the snai2sa24539 allele, genotyping by PCR was sufficient to determine which embryos lacked the wild-type SNP, but not to distinguish wild-type from heterozygote. Thus, images are included for mutants versus “siblings”. For all markers, no obvious defect is detected. Black arrowheads point to the middle of the aortic runx1 or cmyb expression. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. HSC specification was also analyzed in Tg(CD41:GFP/kdrl:mCherry) fish on the snai2112Δ background by confocal microscopy at 48 hpf, and Imaris imaging software was used to remove GFP signal outside of the vasculature (D). Pink coloration is indicative of double positive cells as filtered by the surfaces feature of Imaris. White arrowheads indicate separate putative HSPCs, and quantification for each fish was graphed and statistically analyzed by a non-parametric t-test on Prism (E). Error bars are SEM. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Preliminary investigation of morphant vs. mutant phenotype. WISH within 17 hpf embryos from an in-cross of snai2+/112Δ displayed that the Snail family members snai1a and snai1b are not differentially expressed in mutant embryos as compared to heterozygote and wild-type siblings (A). However, qPCR in pooled embryonic trunks at 26 hpf showed a different trend: all 3 additional members of the Snail family show increased expression (B). This graph presents the average of two independent experiments in which embryonic heads were removed and genotyped, followed by pooling of trunks of the same genotype. Error bars are SD. Snai2 reverse primer is designed within the mutant deletion, so transcript decrease reflects present of mutant transcript. We also observed the effect on emerging HSCs when the SB MO was injected into mutant embryos and their siblings by observing both WISH for runx1 at 26 hpf (C), and the double positive population in Tg(CD41:GFP/kdrl:mCherry) embryos at 48 hpf. Double positive cells were filtered by the surfaces function on Imaris, quantified, and submitted to statistical testing by a non-parametric t-test on Prism (D). Error bars are SEM. By both analyses, HSC specification was affected by SB MO in all genetic backgrounds. Black arrowheads point to the middle of the aortic runx1 expression. **** represents p<0.0001. WT: Wild-type. |
|
Migration of the PLM and vascular cord formation is normal in SB morphants. WISH was performed on embryos injected with SB MO and their uninjected siblings to investigate migration of the PLM and formation of the vascular cord. We analyzed fli1a, a gene actively expressed in the PLM as well as in the fully formed vasculature, as well as kdrl, a marker strong in the fully formed vasculature. At 14 and 18 hpf, fli1a staining showed normal formation of the PLM and timely migration to the midline (A). At 26 hpf, the vascular cord and caudal hematopoietic tissue appear largely normal by both fli1a and kdrl staining; however, the intersomitic vessels seem to have some trouble sprouting dorsally (B). Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. |
|
Primitive hematopoiesis and pronephros formation are unaffected in SB morphants. In order to observe other tissues involved in embryonic hematopoiesis, we assayed primitive hematopoiesis by WISH for the early erythroid marker gata1 (A). SB morphants appeared to have normal primitive hematopoiesis initiation. We also observed formation of the pronephros, which will develop to be the adult HSC niche, by WISH for cdh17 (B). Pronephric formation appeared normal in SB morphants. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. |
|
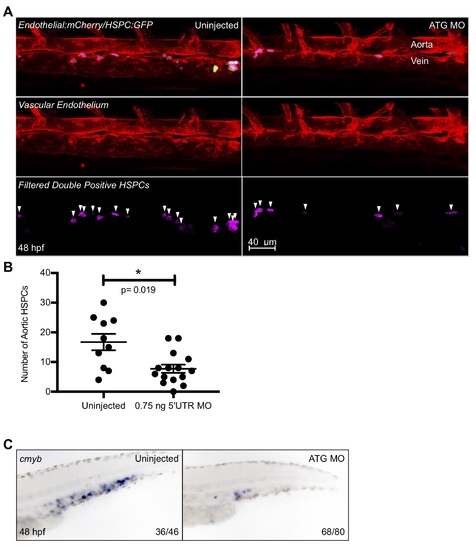
Further analysis of ATG MO hematopoietic phenotype. ATG MO embryos were subjected to WISH for the hematopoietic marker cmyb at 48 hpf (A). The caudal hematopoietic tissue of morphant embryos showed a distinct reduction of cmyb staining as compared to their uninjected siblings. The morpholino was also injected into Tg(CD41:GFP/kdrl:mCherry) embryos and double positive fish were imaged via confocal microscopy at 48 hpf and Imaris imaging software was used to remove GFP signal outside of the vasculature (B). The surfaces feature of Imaris was utilized to quantify double positive cells (shown here in pink), and the resulting data was graphed and statistically analyzed by a non-parametric t-test on Prism (C). Error bars are SEM. There was a small, but significant decrease in the number of HSPCs in the ATG morphant fish. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. |
|
Further notch and somitic morphant data. In order to show not all Notch ligand expression was affected in snai2 SB morphants, we analyzed aortic expression of dll4 and dlc by WISH at 26 hpf (A). SB morphants showed normal levels of both ligands supporting that the aorta is specified correctly. The presence of the Notch intracellular domain in Tg(UAS:NICD-myc) embryos can be assayed by immunohistochemistry for the myc tag, fused to the NICD. Representative images were taken of positive and negative staining present when the transgenic was crossed to the Tg(kdrl:miniGal4) (B). Staining is visible in the dorsal aorta and caudal vein, as well as quite strongly in the caudal hematopoietic tissue of Gal4+/NICD+ embryos. Double fluorescent in situ for dld and myoD was performed in SB morphants and their siblings at 14 hpf, and the results imaged by confocal microscopy (C). Representative images show that morphant embryos have decreased somitic dld staining, especially within the more anterior somites. myoD in the same somites was expressed normally. We also analyzed jam2a expression by WISH (D), since not only is this gene expressed within the somites, but it has been shown to be essential for notch signal transduction to the migrating PLM. SB morphants showed normal expression of jam2a. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. |
|
Snai2112Δ further analysis. A representative gel image shows the different banding pattern observed when genotyping embryos from a snai2+/112Δ in-cross (A). In order to assess later stages of embryonic hematopoiesis, we assessed expression of the T-cell marker, rag1, in 4 dpf embryos (B). Wild-types, heterozygotes, and mutants all showed normal rag1 staining. When snai2 mutants were analyzed on the Tg(CD41:GFP/kdrl:mCherry) background, we simultaneously injected a portion of the clutch analyzed with SB MO. These embryos were imaged via confocal microscopy and Imaris imaging software was used to remove GFP signal outside of the vasculature (C) alongside their uninjected siblings shown in Fig 7D. Quantification is shown in Fig 9D. Additionally, expression of the HSC specification marker, runx1, was analyzed by in situ hybridization at ~26 hpf in embryos injected with snai2 ATG MO, snai1a morpholino (MO), and their siblings. Black arrowheads point to the middle of the aortic runx1 expression. Numbers in the lower right-hand corner of each image depict the number of embryos with the phenotype pictured out of the total number of embryos assayed in each condition. |
|
SB MO causes an increase in p53 transcript, but loss of p53 does not rescue hematopoietic phenotype. The potential of toxicity caused by the SB MO was analyzed by qPCR for p53 in morphant and uninjected pooled embryos at 26 hpf (A). Via this analysis, we saw that indeed p53 transcript was increased as compared to uninjected siblings. We also show snai2 transcript levels, as they are consistently increased in SB MO injected embryos. SB MO was then injected into embryos derived from a p53+/- in-cross, and the embryos analyzed for the hematopoietic markers rag1 and cmyb. The loss of p53 did not appear to rescue the morphant phenotype of decreased levels of both genes. Black arrowheads indicate the general position of the thymus. |