- Title
-
The AP-1 transcription factor component Fosl2 potentiates the rate of myocardial differentiation from the zebrafish second heart field
- Authors
- Jahangiri, L., Sharpe, M., Novikov, N., González-Rosa, J.M., Borikova, A., Nevis, K., Paffett-Lugassy, N., Zhao, L., Adams, M., Guner-Ataman, B., Burns, C.E., Burns, C.G.
- Source
- Full text @ Development
|
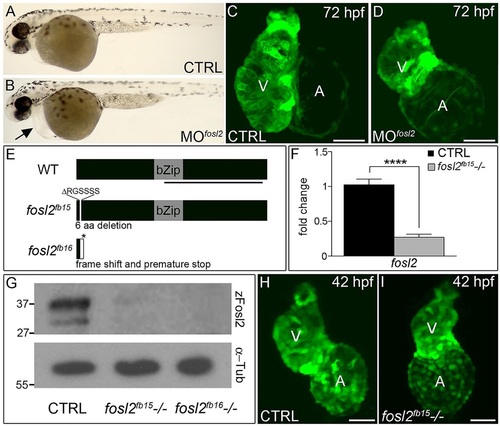
fosl2-null embryos exhibit defects in cardiogenesis. (A,B) Bright-field images of control (CTRL; A; n=44) and fosl2 morphant (MOfosl2; B; n=35) embryos at 72hpf. Morphant embryos exhibit pericardial edema (black arrow). (C,D) Confocal images of GFP+ hearts in 72hpf control (C; n=6) and fosl2 morphant (D; n=9) Tg(cmlc2:GFP) embryos. (E) Schematic diagrams of wild-type (WT) zebrafish Fosl2 with its basic leucine zipper domain (bZip) and the predicted protein products of two fosl2 mutant alleles, fosl2fb15 and fosl2fb16, generated through TALEN-mediated genome editing. The black line highlights the C-terminal region of the protein used to raise polyclonal antiserum. (F) Bar graph showing the relative levels of fosl2 mRNA in control and fosl2fb15-/- embryos at 48hpf as measured by quantitative PCR (n=6 biological replicates per group). Error bars represent s.d. ****P<0.0001. (G) Western blots of 30hpf whole-embryo lysates from control and mutant animals probed with Fosl2 (zFosl2) or α-tubulin (α-Tub) antiserum. (H,I) Confocal images of GFP+ hearts in 42hpf control (H; n=10) and mutant (I; n=4) Tg(cmlc2:GFP) embryos. V, ventricle. A, atrium. Scale bars: 50µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
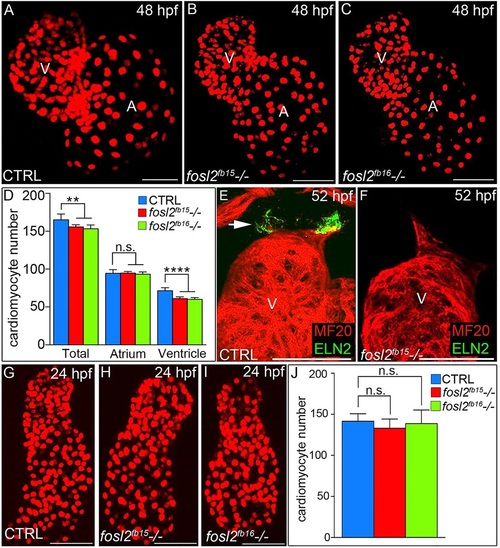
A ventricular cardiomyocyte deficit emerges in fosl2 mutant embryos after unperturbed linear heart tube morphogenesis. (A-C) Confocal images of fluorescent cardiomyocyte nuclei in hearts of 48hpf control sibling (CTRL; A; n=11), fosl2fb15-/- (B; n=5) and fosl2fb16-/- (C; n=5) Tg(cmlc2:DsRed2-nuc) embryos. (D) Bar graph showing mean total, atrial and ventricular cardiomyocyte numbers in each experimental group. Error bars represent s.d. **P<0.01, ****P<0.0001, n.s., not significant. (E,F) Confocal images of ventricular and outflow tract (OFT) regions of 52hpf control (E; n=10) and null mutant (F, n=4) embryos co-stained with antibodies recognizing striated muscle (MF20, red) or OFT smooth muscle (Elastin2, ELN2, green, arrow in E). (G-I) Confocal images of fluorescent cardiomyocyte nuclei in linear heart tubes of 24hpf control (G; n=12), fosl2fb15-/- (H; n=5) and fosl2fb16-/- (I; n=5) Tg(cmlc2:DsRed2-nuc) embryos. (J) Bar graph showing mean cardiomyocyte numbers in each experimental group at 24hpf. Error bars represent s.d. n.s., not significant. V, ventricle. A, atrium. Scale bars: 50µm. |
|
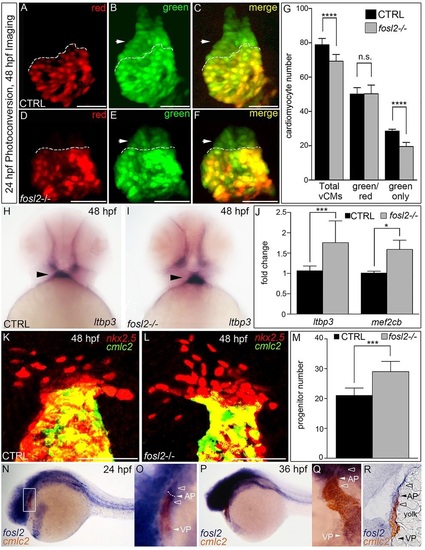
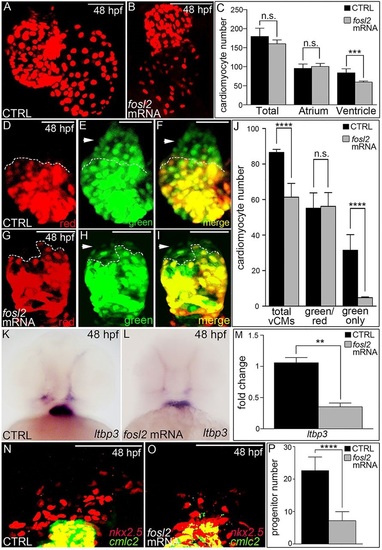
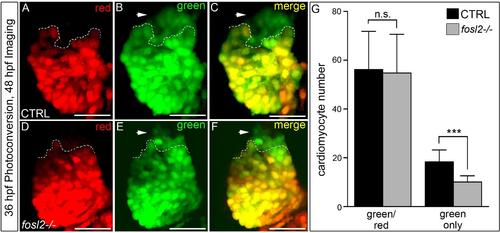
Fosl2 potentiates the progenitor to cardiomyocyte transition during SHF-mediated ventricular growth. (A-G) Cardiomyocyte photoconversion assay. Control sibling (CTRL; A-C; n=23) and fosl2fb16-/- (D-F; n=6) Tg(myl7:nlsKikGR) embryos were photoconverted at 24hpf and imaged by confocal microscopy at 48hpf in the red (A,D) and green (B,E) channels. Merged images are shown in C,F. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads highlight SHF-derived green-only cardiomyocytes. (G) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. ****P<0.0001, n.s., not significant. (H,I) Ventral images of 48hpf control (H; n=16) and fosl2-null (I; n=5) embryos stained with a riboprobe for the SHF marker ltbp3. Arrowheads highlight extra-cardiac SHF progenitors. (J) Bar graph showing relative levels of the SHF markers ltbp3 (n=9 biological replicates per group) and mef2cb (n=3 biological replicates per group) in control and mutant embryos at 48hpf as measured by quantitative PCR. Error bars represent s.d. ***P<0.001, *P<0.05. (K,L) Confocal images of the arterial poles in control (K; n=12) and fosl2 mutant (L; n=4) embryos carrying the Tg(nkx2.5:nZsYellow) and Tg(cmlc2:GFP) transgenes co-stained with antibodies recognizing ZsYellow (red) or GFP (green). (M) Bar graph showing the mean numbers of extra-cardiac SHF progenitor cells (red nuclei without yellow cytoplasm) in both experimental groups. Error bars represent s.d. ***P<0.001. (N-R) Double in situ hybridization analysis of fosl2 (blue) and cmlc2 (red) in whole mounted (N-Q) and sagittally crysectioned (R) 24hpf (N,O) and 36hpf (P-R) embryos. Anterior is to the left in N-P,R. Boxed region in N is magnified in O. (Q) Anterior and dorsal view of the embryo following removal of the head. Open arrowheads in O,Q,R highlight fosl2+ cells on either side of the arterial pole (AP; closed arrowheads and dashed line in O). More than 20 embryos per group were evaluated. VP, venous pole. Scale bars: 50µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
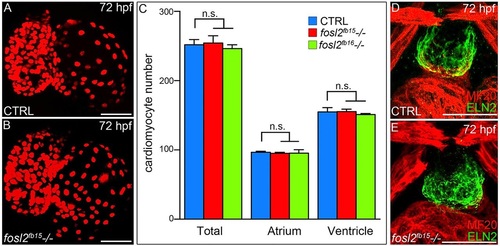
fosl2 mutants recover from their ventricular deficit. (A,B) Confocal images of hearts from 72hpf control sibling (CTRL; A; n=12) and fosl2 (B; n=5) mutant Tg(cmlc2:DsRed2-nuc) embryos. (C) Bar graph showing the mean numbers of total, atrial and ventricular cardiomyocytes in control, fosl2fb15-/- and fosl2fb16-/- (n=3) embryos. Error bars represent s.d. n.s., not significant. (D,E) Confocal images of OFT regions in control (D; n=15) and fosl2 mutant (E; n=4) embryos co-stained with antibodies recognizing striated muscle (MF20, red) or OFT smooth muscle (Elastin2, ELN2, green). Scale bars: 50µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
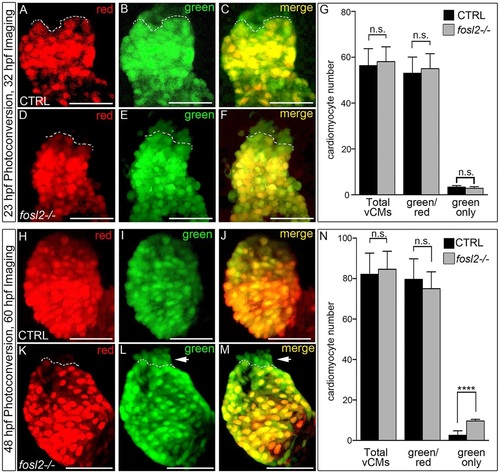
The ventricular deficit resolves in fosl2 mutants through extension of the SHF-mediated cardiomyocyte accretion window. Cardiomyocyte photoconversion assay. Control sibling (CTRL; A-C; n=11) and fosl2-null (D-F; n=7) Tg(myl7:nlsKiKGR) embryos were photoconverted at 23hpf and imaged by confocal microscopy at 32hpf in the red (A,D) and green (B,E) channels. Merged images are shown in C and F. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. (G) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. n.s., not significant. Control (H-J; n=19) and fosl2 mutant (K-M; n=5) Tg(myl7:nlsKiKGR) embryos were photoconverted at 48hpf and imaged by confocal microscopy at 60hpf in the red (H,K) and green (I,J) channels. Merged images are shown in J,M. Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads identify SHF-derived green-only cardiomyocytes that differentiated after 48 hpf specifically in fosl2-null animals. (N) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes and green-only cardiomyocytes. Error bars represent s.d. n.s., not significant, ****P<0.0001. Scale bars: 50µm. |
|
Overexpression of Fosl2 compromises SHF-mediated ventricular growth. (A,B) Confocal images of hearts in 48hpf control sibling (CTRL; A; n=11) and Fosl2-overexpressing (B; n=5) Tg(myl7:nlsDsRed-Express) embryos. (C) Bar graph showing the mean numbers of total, atrial and ventricular cardiomyocytes in both experimental groups. n.s., not significant, ***P<0.001. (D-I) Cardiomyocyte conversion assay. Control (n=7) and Fosl2-overexpressing (n=5) Tg(myl7:nlsKiKGR) embryos were photoconverted at 24hpf and imaged by confocal microscopy at 48hpf in the red (D,G) and green (E,H) channels. Merged images are shown in (F,I). Dashed lines highlight boundaries between cardiomyocytes that differentiated before (bottom) or after (top) photoconversion. Arrowheads identify SHF-derived green-only cardiomyocytes. (J) Bar graph showing the mean numbers of total ventricular cardiomyocytes, green and red positive cardiomyocytes, and green-only cardiomyocytes. Error bars represent s.d. ****P<0.0001, n.s., not significant. (K,L) Ventral images of control (n=30) and Fosl2 (n=35) overexpressing 48hpf embryos stained with a riboprobe for the SHF marker ltbp3. (M) Bar graph showing the relative levels of ltbp3 mRNA at 48hpf in both experimental groups as measured by quantitative PCR (n=3 biological replicates per group). **P<0.01. (N-O) Confocal images of the arterial poles in control (N; n=7) and Fosl2-overexpressing (N; n=6) embryos carrying the Tg(nkx2.5:nZsYellow) and Tg(cmlc2:AmCyan) transgenes co-stained with antibodies recognizing ZsYellow (red) or AmCyan (green). (P) Bar graph showing the mean numbers of extra cardiac SHF progenitor cells (red nuclei without yellow cytoplasm) in both experimental groups at 48hpf. Error bars represent s.d. ****P<0.0001. Scale bars: 50µm. EXPRESSION / LABELING:
|
|
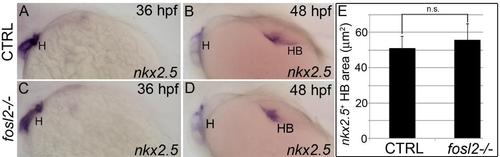
Hepatobiliary morphogenesis proceeds at a normal rate between 36 hpf and 48 hpf in fosl2 mutants. (A-D) In situ hybridization analysis of nkx2.5 transcripts in control sibling (CTRL; A,B) and fosl2- /- (C,D) embryos at 36 hours post fertilization (hpf) (A, C; n=>9 embryos) and 48 hpf (B,D). (E) Bar graph showing the average sizes of the nkx2.5 signal in 48 hpf control (n=18) and mutant (n=26) embryos. Error bars represent one standard deviation. n.s., not significant. H, heart. HB, hepatobiliary system. |
|
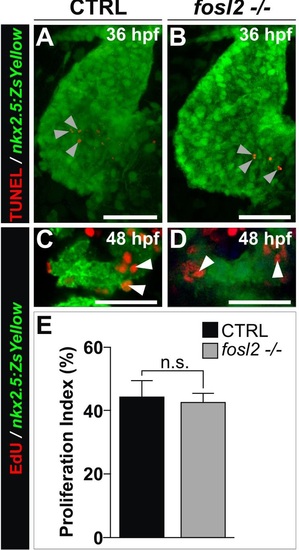
Analysis of proliferation and apoptosis in fosl2 mutants. (A,B) Confocal images of 36 hours post-fertilization (hpf) control sibling (CTRL; A; n=4) and fosl2-/- (B; n=4) Tg(nkx2.5:ZsYellow) embryos processed for the detection of apoptotic cells (red) using the TUNEL assay. Embryos were coimmunostained to detect the ZsYellow fluorescent protein (green). Although apoptotic cells were detected within the Z-stacks (arrowheads), they were never found overlapping ZsYellow+ myocardium in either experimental group. (C,D) Confocal images of the ZsYellow+ SHF in 48 hpf control (C; n=4) and fosl2fb16-/- (D; n=4) Tg(nkx2.5:ZsYellow) embryos double immunostained for ZsYellow (green) and EdU (red) following exposure to EdU at 36 hpf for 30 minutes. Embryos were co-stained with DAPI (not shown) to visualize all nuclei. (E) Bar graph showing the average percentages of DAPI+, ZsYellow+ cells that were also EdU+. Error bars represent one standard devision. n.s., not significant. Scale bars: 50µm in (A,B) and 25µm in (C,D). |
|
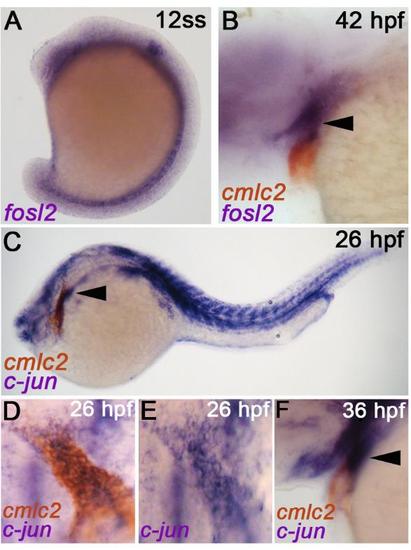
Developmental expression profiles of AP-1 components fosl2, c-jun, and c-fos. (A, B) In situ hybridization analysis of fosl2 transcripts at the 12 somites stage (ss) (A) and 42 hours post fertilization (hpf) (B). The embryo shown in (B) is co-stained for the myocardial transcript cmlc2. The arrowhead in (B) highlights fosl2 expression in pharyngeal mesoderm on the extra-cardiac side of the arterial pole where SHF progenitors reside. (C-F) Double in situ hybridization analysis of c-jun and cmlc2 transcripts at 26 hpf (C-E) and 36 hpf (F). Double stained embryos shown in (D) were washed with methanol to remove the cmlc2 signal and photographed again (E). C-jun+ cells were observed on either side of the arterial pole. Arrowhead in (F) highlights c-jun expression in pharyngeal mesoderm on the extra cardiac side of the arterial pole where SHF progenitors reside. In all cases, greater than 20 embryos were analyzed. |
|
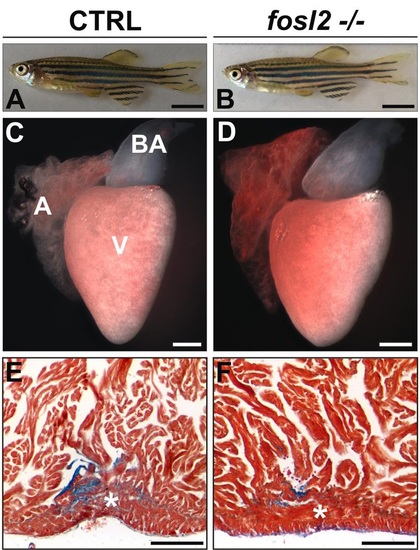
fosl2-/- adults are indistinguishable from siblings with regard to animal size, heart size, heart morphology, and cardiac regenerative capacity. (A,B) Photographs of 6-month old sibling control (CTRL; A; n=6) and fosl2 null (B; n=6) zebrafish showing their equivalent sizes. (C,D) Photographs of hearts dissected from 6 month old control (C; n=6) and fosl2 null (D, n=6) zebrafish showing their equivalent sizes and morphologies. (E,F) AFOG-stained cardiac sections from 6 month-old control (E; n=4 hearts) and fosl2 null (F; n=4 hearts) hearts on day 45 post apical resection. Mutant hearts regenerated (*) resected myocardium normally. A, atrium. V, ventricle. BA, bulbus arteriosus. Scale bars: 1mm in (A,B), 200µm in (C-F). |
|
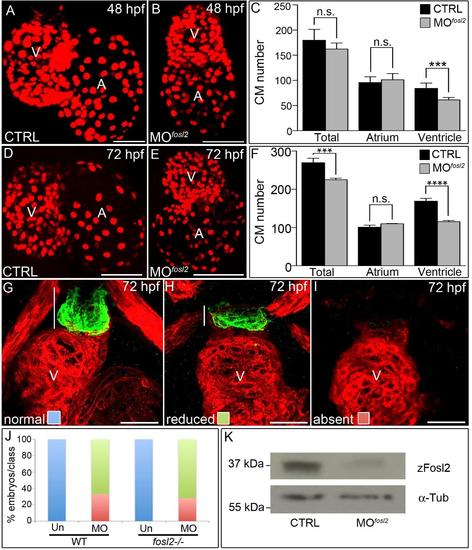
Analysis of fosl2 morphants. (A,B,D,E) Confocal images of hearts in 48 hpf (A,B) and 72 hpf (D,E) control sibling (CTRL; A; n=7 and D; n=12) and fosl2 morphant (B; n=6 and E; n=7) Tg(cmlc2:dsRed2-nuc) embryos. (C,F) Bar graphs showing average total, atrial, and ventricular cardiomyocyte numbers in each experimental group at 48 hpf (C) and 72 hpf (F). Error bars represent one standard deviation. n.s., not significant. ***p<0.001. (G-I) Confocal images of ventricular and outflow tract (OFT) regions of 72 hpf control (G) and fosl2 morphant (H, I) embryos co-stained with antibodies recognizing striated muscle (red) or Eln2+ OFT smooth muscle (green) that demonstrate normal (G), reduced (H), or absent (I) OFT Eln2. White lines approximate the sizes of the OFTs. (J) Bar graph showing the percentages of morphant (WT embryos injected with the fosl2 morpholino; n=18 total) and morphant-mutant (fosl2-/- embryos injected with the morpholino; n=25 total) embryos that fall into each phenotypic class shown in (G-I). Uninjected (Un) WT (n=22) and fosl2-/- embryos (n=22) were used as controls. (K) Western blots of 30 hpf whole-embryo lysates from control and morphant embryos probed with Fosl2 (zFosl2) or α-Tubulin (α-Tub) antiserum. V, ventricle, A, atrium. |
|
Analysis of SHF-mediated cardiomyocyte accretion between 36 and 48 hours post-fertilization in fosl2-/- embryos. (A-G) Cardiomyocyte conversion assay. Control sibling (n=10) and fosl2-/- (n=11) Tg(myl7:nlsKiKGR) embryos were photoconverted at 36 hours post fertilization (hpf) and imaged by confocal microscopy at 48 hpf in the red (A, B) and green (B, E) channels. Merged images are shown in (C, F). (G) Bar graph showing the average numbers of green and red positive ventricular cardiomyocytes and green only cardiomyocytes. Error bars represent one standard deviation. ***p<0.001. n.s., not significant. |
|
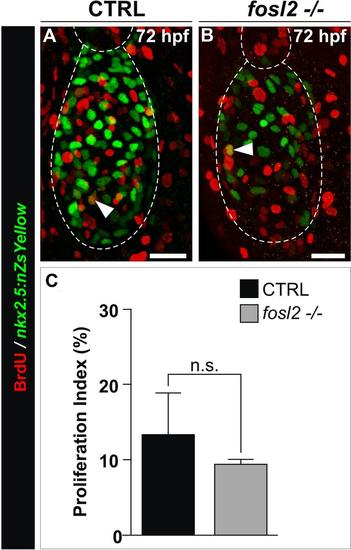
Analysis of ventricular cardiomyocyte proliferation in fosl2-/- animals. (A,B) Confocal images of 72 hpf control sibling (CTRL; A, n=5) and fosl2fb16-/- (B; n=4) Tg(nkx2.5:ZsYellow-nuc) embryos exposed to BrdU between 48 hpf and 72 hpf and double immunostained for ZsYellow protein (green) and BrdU (red). The arrows highlight double positive nuclei. (C) Bar graph showing the average percentages of ZsYellow+ ventricular nuclei that were BrdU+. Error bars represent one standard deviation. n.s., not significant. Scale Bars: 25µm. |
|
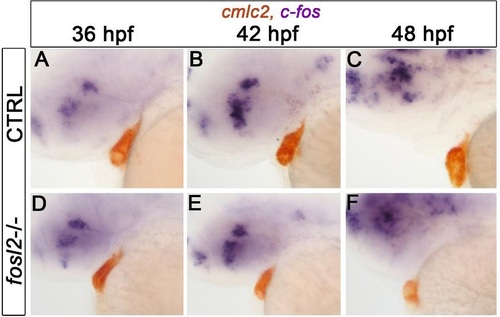
In situ hybridization analysis of c-fos. Double in situ hybridization analysis of c-fos and cmlc2 transcripts at 36 hpf (A), 42 hpf (B) and 48 hpf (C). Greater than 20 embryos were evaluated at each developmental stage. |