- Title
-
Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Authors
- Sasaki, T., Lian, S., Qi, J., Bayliss, P.E., Carr, C.E., Johnson, J.L., Guha, S., Kobler, P., Catz, S.D., Gill, M., Jia, K., Klionsky, D.J., Kishi, S.
- Source
- Full text @ PLoS Genet.
|
Aberrant autophagosome and autolysosome formation in spns1-mutant zebrafish. (A) Yolk opaqueness and LC3 puncta formation in spns1-mutant zebrafish embryos. For EGFP-LC3 transgenic spns1-mutant [Tg(CMV:EGFP-LC3);spns1hi891/hi891] fish siblings, bright-field and fluorescence images of wild-type (wt) control (upper) and spns1 mutant (spns1-/-) (lower) embryos at 84 hpf are shown. The black arrow indicates the yolk-opaqueness phenotype in the spns1 mutant. The gross expression of EGFP-LC3 at head and trunk in the spns1-mutant animal is relatively stronger than in the wt animal. Occasionally, however, a high intensity signal can be observed at the liver region in the mutant (as seen in D). Scale bar, 250 µm. (B) EGFP-LC3 punctate compartments in the liver cells of the spns1 mutant. Through high magnification (×600) confocal microscopy, intracellular EGFP-LC3 puncta were visualized in live animals at 84 hpf. Nuclei were counterstained with Hoechest 33342 (blue), and peri-nuclear EGFP-LC3 puncta were evident in the spns1 mutant, but not in wt animals. Scale bar, 10 μm. (C) Immunoblotting to detect the conversion of LC3-I to LC-II. Using an anti-LC3 antibody, both endogenous LC3 and transgenic (exogenous) EGFP-LC3 expression was detected and an increase of LC3-II conversion/accumulation was seen in the spns1 mutant compared with wt fish at 84 hpf. (D–F) Identification of autophagosome and autolysosome/lysosome formation in the spns1 mutant. (D, E) LysoTracker (DND-99; red) staining of EGFP-LC3 transgenic spns1-mutant [Tg(CMV:EGFP-LC3); spns1hi891/hi891] embryos was performed at 84 hpf. At the whole animal levels (D), the EGFP-LC3 signal is relatively higher throughout in the spns1 mutant than in wild type, and a particularly strong signal can be seen in the liver, as shown in (A). In the head and trunk portions of the animals (D), a distinctive increase in the intensity of LysoTracker can be observed in the spns1 mutant. At the intracellular level (E), several small LC3 spots and largely diffuse green signal in the cells and cytosolic LysoTracker staining is seen. A number of enlarged LC3- and LysoTracker-positive yellow punctate structures can be seen in the spns1 mutant by confocal microscopy at a higher magnification (inset; enlarged from dotted square area). (F) EGFP-LC3 and mCherry-LC3 double-transgenic [Tg(EGFP-LC3:mCherry-LC3)] zebrafish were used to monitor autolysosome formation in spns1 MO-injected embryos at 84 hpf. A number of enlarged yellow LC3 puncta were detected in the spns1 morphant, while only small yellow LC3 spots can be seen in control-injected embryos. Nuclei were counterstained with 42, 6-diamidino-2-phenylindole, dihydrochloride (DAPI). Scale bar, 250 μm in (D). Scale bar, 10 μm in (E, F). Quantification of data presented in D (n = 12), E (n = 6), and F (n = 6) is shown in the right graph; the number (n) of animals is for each genotype. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (G) Transgenic expression of mCherry-Lamp1 in wt [Tg(CMV:EGFP-LC3)] and spns1-mutant [Tg(CMV:EGFP-LC3);spns1hi891/hi891] animals 84 hpf. Scale bar, 10 µm. (H) Transgenic expression of EGFP-Vector (vector), EGFP-wild-type Spns1 (spns1 WT), or EGFP-mutant Spns1 (spns1 E153K) in [Tg(CMV:mCherry-LC3);spns1hi891/hi891] animals at 84 hpf. Scale bar, 10 μm. Quantification of data presented in H is shown for ratio of yolk opaqueness phenotype (n = 48), mCherry intensity (red) (n = 6), and merge intensity of EGFP and mCherry (yellow) (n = 6) in the right graphs; the number (n) of animals is for each genotype. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± standard deviation (S.D.), *p<0.005; ns, not significant. EXPRESSION / LABELING:
PHENOTYPE:
|
|
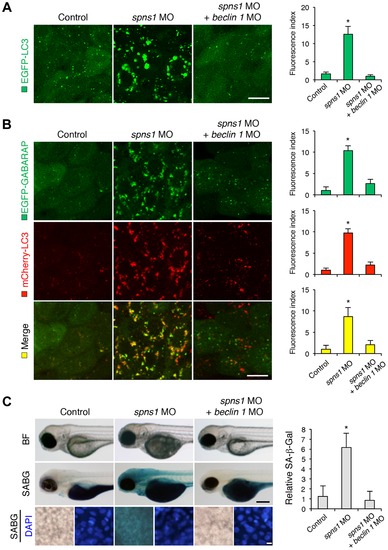
Knockdown of beclin 1 suppresses the Spns1 deficiency in zebrafish. (A) Schematic representation of the zebrafish beclin 1 (zbeclin 1) gene, its mRNA and protein products. A splice-blocking beclin 1 MO was designed to overlap the intron-exon boundary at the 52-splice junction of exon 4 in the zebrafish beclin 1 gene. To detect aberrantly spliced RNA products, two forward primers were designed for exon 3 (EX3 primer) and exon 4 (EX4 primer), and one reverse primer was designed for exon 7 (EX7 primer) within the beclin 1 gene. The zebrafish beclin 1 gene has a total of 11 exons having three unique domains [BH3 domain, coiled-coil (CCD) domain, and evolutionarily conserved (ECD) domain], and the beclin 1 MO was anticipated to disrupt the BH3 domain encoded by exon 4 and exon 5. (B) Splicing detection of zbeclin 1 mRNA by RT-PCR. Amplified PCR fragments show the intact sizes of the two amplicons for EX3-EX7 and EX4-EX7 following control (water) injection or only spns1 MO injection. Either beclin 1 MO (12 ng/embryo) injection or coinjection of spns1 MO (4 ng/embryo) and beclin 1 MO (12 ng/embryo) generated a skipping of exon 4 (beclin 1Δexon4). This was detected by the presence of an altered EX3-EX7 amplicon and undetectable EX4-EX7 product. The deletion of exon 4 was confirmed by sequencing. Injected embryos were harvested for total RNA isolation at 54 hpf. (C and D) Rescue of the spns1 morphant by beclin 1 knockdown. (C) The yolk opaqueness phenotype appearance in control-injected (water), spns1 MO-injected, and spns1 and beclin 1 MOs-coinjected embryos was followed through 72 hpf. At 24 hpf, opaqueness commenced from the yolk extension region, which had almost disappeared or was severely damaged (more than 95% of spns1 MO-injected animals) with an extension of opacity to the entire yolk at 48 hpf. By 72 hpf, yolk opaqueness became highly dense throughout most of the spns1 MO-injected embryos, which usually died within another 24 h. Scale bar, 250 μm. (D) Clarification of the yolk opaqueness phenotype in spns1 morphants at 72 hpf. As described in (C), more than 95% of the spns1 MO-injected embryos showed a ‘mostly opaque’ yolk at 48 hpf, and such embryos subsequently died. Animals showing a ‘partially opaque’ yolk could sometimes be recovered and subsequently survived 96 h and beyond. beclin 1 MO coinjection dramatically increased (more than 10 times) the animal numbers with the partial yolk opaque phenotype. (E) Survival curve for spns1 morphant and spns1;beclin 1-double morphant larvae (log rank test: χ2 = 162.5 on one degree of freedom; p<0.0001). |
|
Knockdown of beclin 1 suppresses abnormal autolysosomal puncta formation and embryonic senescence caused by Spns1 deficiency in zebrafish. (A) Effect of beclin 1 knockdown on EGFP-LC3 puncta formation in spns1-depleted zebrafish embryos. Injection of control (water) injection, spns1 MO (4 ng/embryo) or coinjection of spns1 MO (4 ng/embryo) and beclin 1 MO (12 ng/embryo) into Tg(CMV:EGFP-LC3) fish was performed to assess whether the beclin 1 knockdown reduces or eliminates aggregated LC3 puncta induced by Spns1 depletion at 84 hpf. Scale bar, 10 μm. Quantification of data presented in panel A (n = 12) is shown in the right graph; the number (n) of animals is for each morphant or water-injected control. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (B) Effect of beclin 1 knockdown on EGFP-GABARAP as well as mCherry-LC3 puncta formation in spns1-depleted zebrafish embryos. Injection of control (water), spns1 MO or coinjection of spns1 MO and beclin 1 MO into Tg(CMV:EGFP-GABARAP;mCherry-LC3) fish was performed to evaluate whether the beclin 1 knockdown reduces or eliminates the aggregation of GFP-GABARAP puncta in comparison with those of LC3 caused by the Spns1 depletion at 84 hpf. Scale bar, 10 μm. Quantification of data presented in the top row (green; EGFP) (n = 9), middle row (red; mCherry) (n = 12), and bottom row (yellow; merge of EGFP and mCherry) (n = 9) in panel B is shown in the right graphs; the number (n) of animals is for each morphant or water-injected control. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (C) Effect of beclin 1 knockdown on embryonic senescence in spns1 morphant. By using the same injection samples [injection of control (water), spns1 MO or coinjection of spns1 MO and beclin 1 MO into Tg(CMV:EGFP-GABARAP;mCherry-LC3) fish], SA-β-gal staining was performed to assess whether the beclin 1 knockdown has any impact on the embryonic senescence caused by Spns1 depletion at 84 hpf. Representative images of individual fish by bright field (BF, live samples) and SA-β-gal (SABG) staining are shown in the upper and middle panels, respectively. Scale bar, 250 μm. Lower panels are larger magnification images of corresponding SA-β-gal samples shown in the middle panels and the fluorescence images of nuclei counterstained with DAPI. Scale bar, 10 μm. Quantification of data presented in the middle row (SABG) in panel C (n = 12) is shown in the right graph; the number (n) of animals is for each morphant or water-injected control. Error bars represent the mean ± S.D., *p<0.005. EXPRESSION / LABELING:
PHENOTYPE:
|
|
p53 depletion does not suppress but rather exacerbates Spns1 deficiency. (A) Effect of p53 knockdown on embryonic senescence and autolysosome formation in spns1 morphants. The impact of transient p53 knockdown on SA-β-gal (SABG) induction, as well as on EGFP-LC3 and LysoTracker (LysoT) puncta, was determined in spns1 morphants at 84 hpf, followed by the MO (4 ng/embryo) injections. Inverse-sequence p53 MO (inv. p53 MO) was used as a negative control for the original p53 MO. Scale bar, 250 μm in the SABG images. Scale bar, 10 μm in the fluorescence images. (B) Quantification of the SA-β-gal intensities in MO-injected animals, as shown for the SABG images in (A). Quantification of data presented in the top row (SABG) in B (n = 12) is shown; the number (n) of animals is for each morphant. (C) Quantification of EGFP-LC3 and LysoTracker puncta in MO-injected animals shown in (A) (n = 9); the number (n) of animals is for each morphant. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (D) Effect of a p53 mutation on embryonic SA-β-gal activity in the spns1 mutant. The heritable impact of p53 and Spns1 on SA-β-gal induction was tested in each single gene mutant [spns1hi891/hi891 (spns1-/-) or tp53zdf1/zdf1 (p53m/m)] and double mutant spns1hi891/hi891;tp53zdf1/zdf1 (spns1-/-;p53m/m) compared with wild-type (wt) animals at 84 hpf. Scale bar, 250 μm. (E) Quantification of the SA-β-gal intensities in wt, tp53zdf1/zdf1, spns1hi891/hi891 and spns1hi891/hi891;tp53zdf1/zdf1 animals, shown in (D). Quantification of data presented in panel D (n = 12) is shown; the number (n) of animals is for each genotype. (F) Quantitative RT-PCR analyses of senescence marker and/or mediator expression as well as p53-downstream target genes in wt, tp53zdf1/zdf1, spns1hi891/hi891 and spns1hi891/hi891;tp53zdf1/zdf1 at 72 hpf. Data are mean ±SD [n = 4 samples (3 embryos/sample) per genotype]. Asterisks denote significant changes compared to wt values. *p<0.05. (G) LC3 conversions in p53 and spns1-mutant animals. Protein detection for the conversion/accumulation of LC3-I to LC-II was performed in the described mutant background animals in comparison with wt fish at 84 hpf. Western blot analysis using anti-LC3 antibody shows endogenous LC3 protein levels, which can confirm an increase of the total amount of LC3 in the p53 mutant compared with wt fish. Increased LC3-II conversion/accumulation was detected in p53 and spns1 double-mutants as well as in spns1 single-mutant fish. (H) The blotting band intensities of LC3-I, LC3-II and β-actin were quantified (n = 6), and the relative ratios between LC3-II/actin and LC3-I/actin are shown in the bar graph; the number (n) of animals is for each genotype. (I) wt, tp53zdf1/zdf1, spns1hi891/hi891 and spns1hi891/hi891;tp53zdf1/zdf1 embryos injected with beclin 1 MO or control MO (12 ng/embryo) were assayed for SA-β-gal at 84 hpf. beclin 1 MO-mediated suppression of SA-β-gal in spns1hi891/hi891 animals was attenuated in the p53 mutant background. Scale bar, 250 μm. (J) Quantification of the SA-β-gal intensities shown in (I). Quantification of data presented in H (n = 12) is shown; the number (n) of animals is for each genotype with MO. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Acidity-dependent lysosomal biogenesis is rate limiting in spns1-mutant zebrafish. (A) Effect of bafilomycin A1 (BafA) on the yolk opaque phenotype (BF; bright field) and embryonic senescence (SABG; SA-β-gal) in the spns1 mutant in the presence or absence of p53 at 48 hpf. Normal wild-type (spns1+/+;p53+/+), tp53zdf1/zdf1 (p53m/m), spns1hi891/hi891 (spns1-/-) and spns1hi891/hi891;tp53zdf1/zdf1 (spns1-/-;p53m/m) embryos at 36 hpf were incubated with BafA (200 nM) for 12 h, and stained with LysoTracker at 48 hpf, followed by SA-β-gal staining at 60 hpf. Scale bar, 250 μm. (B) Quantification of the SA-β-gal intensities shown in (A). Quantification of data presented in panel A (n = 12) is shown; the number (n) of animals is for each genotype with DMSO or BafA. (C) Gross morphology, EGFP-LC3 and LysoTracker intensities in wild-type (wt) and spns1-mutant animals treated with BafA shown at 48 hpf (12 h treatment starting at 36 hpf). Scale bar, 250 μm. (D) Quantification of the EGFP-LC3 and LysoTracker fluorescence intensities shown in (C). Quantification of data presented in the middle and bottom rows (green; EGFP, red; mCherry) in panel C (n = 12) is shown; the number (n) of animals is for each genotype with DMSO or BafA. (E) Intracellular autolysosome formation and lysosomal biogenesis in the BafA-treated spns1 mutant. The samples analyzed in (C) were observed by using confocal microscopy at high magnification (×600). Scale bar, 10 μm. (F) Quantification of the EGFP-LC3 and LysoTracker fluorescence intensities shown in (E). Quantification of data presented for EGFP (green) and mCherry (red) signals in panel E (n = 6) is shown; the number (n) of animals is for each genotype with DMSO or BafA. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (G) Insufficient intracellular acidity constituent in the spns1 mutants. Using two different acidic-sensitive probes, LysoSensor 189 and neutral-sensitive LysoSensor 153 (green), in combination with LysoTracker (red), wt and spns1-mutant animals showed detectable signals when stained at 72 hpf. In spns1-mutant animals, autolysosomal and/or lysosomal compartments were more prominently detectable by LysoSensor 153 than by LysoSensor 189, at the cellular level with enhanced signal intensity of these enlarged compartments. In stark contrast, the cellular compartments in wt fish treated with pepstatin A and E-64-d (P/E) (12 h treatment from 60 hpf through 72 hpf) were more prominently detectable by LysoSensor 189 than by LysoSensor 153 under the identical LysoTracker staining conditions. Of note, these autolysosomal and lysosomal compartments in spns1 mutants, as well as in wt animals treated with pepstatin A and E-64-d, may still retain some weak (higher pH) and strong (lower pH) acidity, respectively, as short-term BafA treatment (for 1 h between 71 and 72 hpf) can abolish the acidic compartments stained by both LysoSensor and LysoTracker (Figure S17C and D). Scale bar, 10 μm. (H) Quantification of the LysoSensor (189 and 153) and LysoTracker fluorescence intensities shown in (G). Quantification of data presented for LysoSensor (green) and LysoTracker (red) signals in panel G (n = 6) is shown; the number (n) of animals is for each genotype with DMSO or pepstatin A and E-64-d (P/E). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
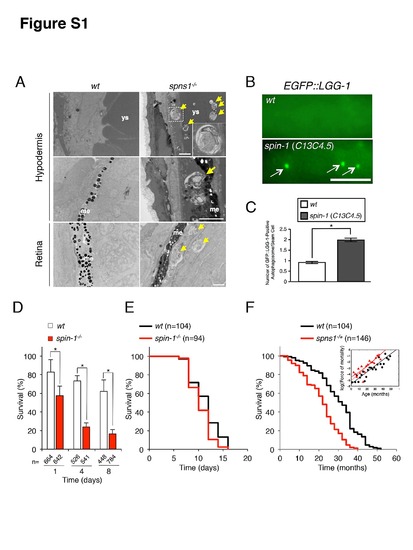
Autophagic abnormalities and survival in spns1-mutant fish and worms. (A) Representative transmission electron microscopy images of normal wt or spns1-mutant fish larvae at 84 hpf. Compared with wild-type (wt) control (left), the spns1 mutant (spns1-/-) (right) accumulates abnormal cytoplasmic inclusions at the hypodermal regions adjacent to yolk sac (ys) (upper two panels) or melanophores (me) (middle two panels), and in the retinal pigment epithelium containing melanophores (me) (lower two panels). Arrows indicate cytoplasmic membranous inclusions. In the right-upper panel, the inset shows a magnified image of the cytoplasmic inclusion surrounded with a dotted square. Scale bar, 2 μm. (B) Modulation of autophagy activity by a mutation in the spns1 homolog (spin-1-/-; C13C4.5) in C. elegans. Representative images of autophagosomes (EGFP::LGG-1 puncta) in seam cells are shown for wild-type {wt, adIs2122 [lgg-1p::GFP::lgg-1, rol-6(su1006)]} animals and for nematodes carrying a homozygous spin-1 deletion allele {spin-1(ok2087); adIs2122 [lgg-1p::GFP::lgg-1+rol-6(su1006)}. Arrows indicate autophagosomes only in the spin-1-/- animals. Scale bar, 5 μm. (C) Quantification of EGFP::LGG-1 puncta is shown for the indicated genetic backgrounds and conditions. The count of puncta per seam cell was 0.8936±0.0926 for wt and 1.9899±0.1396 for spin-1(ok2087) L4 larva, respectively [values are the mean ± standard error of mean (S.E.M.) for 94 (wt) and 99 [spin-1(ok2087)] seam cells; more than 20 animals were examined for each strain] (t-test: *p<0.0001). (D) The starvation sensitivity in spin-1(ok2087) mutant worms. Percent of worms surviving to adulthood on NGM plates with OP50 bacteria after incubation in M9 buffer in the absence of food at the L1 larval stage for the indicated times. Error bars are for standard errors of means estimated assuming a Poisson distribution, and similar results were obtained in three independent experiments. (E) Lifespan in spin-1(ok2087) mutant worms is demonstrated by Kaplan-Meier survival analysis. spin-1(ok2087) mutant worms are short lived compared with the wild-type N2 strain on HT115 bacteria. The median lifespan was 12 days for N2 and 10 days for spin-1(ok2087) (log rank test: χ2 = 8.834 on one degree of freedom; p = 0.003). Similar results were obtained in 2 experiments with 3 independent replicates each. (F) Shorter lifespan in heterozygous spns1-/+ adult fish is demonstrated by Kaplan-Meier survival analysis (log rank test: χ2 = 54.05 on one degree of freedom; p<0.0001) and validated by Gompertz-Makeham model. |
|
Detection of lysosomal and mitochondrial biogenesis in spns1-mutant animals. (A) Whole-mount double staining of live embryos with LysoTracker (10 μM, DND-99; red) and MitoTracker (1 μM, green) at 72 hpf. Intense LysoTracker staining was detected only in spns1 mutants but not in wt animals. In contrast, MitoTracker detected equivalent signals between wt and spns1-mutant animals. Scale bar, 250 µm. (B) Whole-mount double staining of live embryos with MitoSox (5 μM, red) and MitoTracker (1 μM, green) at 72 hpf. Both of the probes detected equivalent signals between wt and spns1-mutant animals. Scale bar (black) in large image, 250 μm. Scale bar (white) in inset, 10 μm. (C) Acidity-dependent quenching of EGFP-LC3 at the LysoTracker-positive compartments in the cells from pepstatin A (5 μg/ml)- and E-64-d (5 μg/ml)-co-treated (P/E) zebrafish embryos at 72 hpf. Scale bar, 10 μm. (D) Acidity-dependent quenching of EGFP-LC3, but not mCherry-LC3, in cells from pepstatin A (5 μg/ml)- and E-64-d (5 μg/ml)-co-treated (P/E) zebrafish embryos at 72 hpf. Scale bar, 10 μm. (E) The degradation capacity of autolysosomes and lysosomes was examined by injection of a lysosomal substrate, DQ Red BSA (DQ-BSA; red) at 60 hpf. The enzyme-catalyzed hydrolysis of the intramolecular self-quenched DQ Red BSA by lysosomal proteases relieves the self-quenching, yielding brightly fluorescent reaction products. DQ Red BSA-injected wt control or spns1-mutant fish expressing EGFP-LC3 were observed at the cellular level by confocal microscopy. Scale bar, 10 μm. Quantification of data presented in E (n = 6), is shown in the right graph; the number (n) of animals is for each genotype. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± standard deviation (S.D.), *p<0.005. |
|
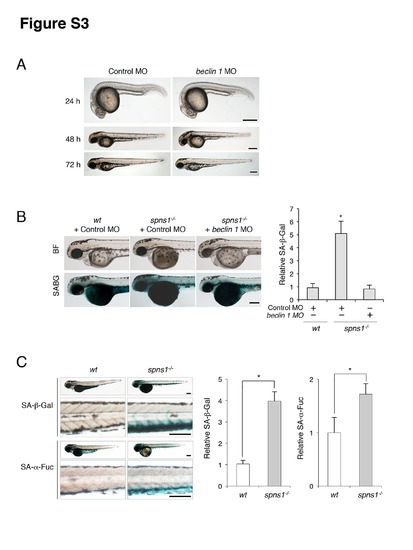
Impact of Beclin 1 depletion on the yolk opaque phenotype and embryonic senescence in spns1-mutant zebrafish. (A) Phenotype of beclin 1 morphant (beclin 1 MO, 12 ng/embryo) at 24, 48 and 72 hpf. Scale bar, 250 μm. (B) Effect of beclin 1 knockdown in the spns1 mutant on the phenotypes of yolk opacity (BF; bright field) and on embryonic senescence (SABG; SA-β-gal) in the spns1 mutant. Following injection of standard control MO or beclin 1 MO (12 ng/embryo) into Tg(CMV:EGFP-LC3); spns1hi891/hi891 embryos, SA-β-gal staining was performed to determine whether the beclin 1 knockdown had any impact on embryonic senescence caused by Spns1 depletion at 84 hpf. Scale bar, 250 μm. Quantification of data presented in panel B (n = 12) is shown in the right graph; the number (n) of animals is for each morphant. (C) Parallel analyses of SA-β-gal and SA-α-fuc demonstrate the significant inductions of both activities in spns1-mutant animals at 84 hpf. As shown in the magnified panels, the caudal venous plexus (CVP) was the most prominently stained region. Staining for SA-β-gal was more intensive than for SA-α-fuc. Scale bar, 250 μm. Quantification of data presented in panel C (n = 12) is shown in the right graph; the number (n) of animals is for each morphant. Error bars represent the mean ± S.D., *p<0.005. |
|
Effect of UV irradiation on spns1-mutant zebrafish. (A) Acrdine orange (green) and Lysotracker (red) intensities, as well as gross morphology, in wild-type (wt) and spns1-mutant animals treated with UV. The UV (18 mj/cm2) treatment was done at 36 hpf, and phenotypes were observed at 48 hpf. Scale bar, 250 μm. Quantification of data presented in A (n = 9) is shown in the right graphs; the number (n) of animals is for each genotype with or without UV treatment. (B) Cellular characteristics in the animals shown in (A) were observed by using confocal microscopy at high magnification (×600). Scale bar, 10 µm. Quantification of data presented in B (n = 6) is shown in the right graphs; the number (n) of animals is for each genotype with or without UV treatment. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
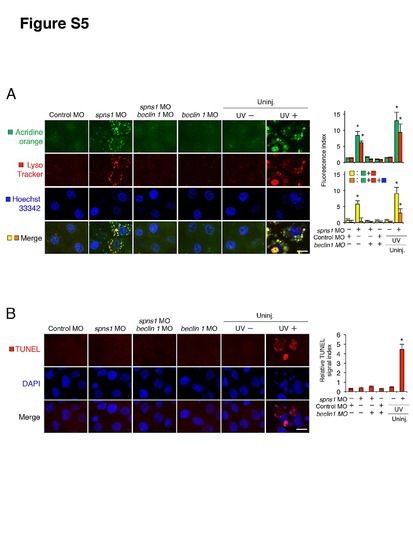
Undetectable apoptosis in spns1 and/or beclin 1 morphants. (A) In spns1 and/or beclin 1 morphants stained with acridine orange (green) and LysoTracker (red) cellular characteristics were compared with UV-treated specimens by using confocal microscopy at high magnification (×600). Scale bar, 10 μm. Quantification of data presented in A (n = 6) is shown in the right graphs; the number (n) of animals is for each morphant and uninjected (Uninj.) animal with or without UV treatment. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (B) TUNEL assays demonstrate apoptosis induction in UV-treated zebrafish embryos, but not in spns1 and/or beclin 1 morphants. The UV (18 mj/cm2) treatment was done at 36 hpf, and phenotypes were observed at 48 hpf. Scale bar, 10 μm. Quantification of the fluorescence intensities is shown at the right-side graph. Quantification of data presented in B (n = 6) is shown in the right graphs; the number (n) of animals is for each morphant and uninjected (Uninj.) animal with or without UV treatment. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005. |
|
Impact of the beclin 1 knockdown on UV-induced apoptosis and autophagy. (A) Partial but significant suppression of UV-induced apoptosis in beclin 1 morphants. The UV (18 mj/cm2) treatment was done at 66 hpf, followed by the phenotype observations at 72 hpf. Scale bar in the large image, 250 µm. Scale bar in the inset, 10 μm. Quantification of data presented in A (n = 9) is shown in the right graphs; the number (n) of animals is for each morphant with or without UV treatment. Three independent areas (periderm or basal epidermal cells in the caudal fin) were selected from individual animals. (B) Sufficient suppression of UV-induced autophagy in beclin 1 morphants. The UV (18 mj/cm2) treatment was done at 69 hpf, followed by the phenotype observations at 72 hpf. Scale bar, 10 μm. Quantification of data presented in A (n = 9) is shown in the right graphs; the number (n) of animals is for each morphant with or without UV treatment. Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., **p<0.005; *p<0.05 in (A), and *p<0.005; ns, not significant in (B). |
|
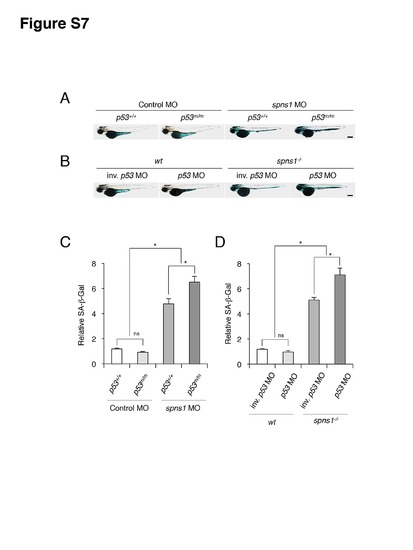
Effects of spns1 and p53 knockdowns on embryonic SA-β-gal activity in p53 and spns1 mutants, respectively. (A) Effect of spns1 knockdown on embryonic senescence in p53 mutants. The impact of transient spns1 knockdown on SA-β-gal induction was determined in spns1 MO-injected tp53zdf1/zdf1 animals at 72 hpf. Standard control MO was used for control injections. Scale bar, 250 μm. (B) Effect of p53 knockdown on embryonic senescence in spns1 mutants. The impact of transient p53 knockdown on SA-β-gal induction was determined in p53 MO-injected spns1hi891/hi891 animals at 72 hpf, followed by the MO injections. Inverse p53 MO (inv. p53 MO) was used for control injections. Scale bar, 10 µm. (C) Quantification of the SA-β-gal intensities shown in (A). Quantification of data presented in panel A (n = 12) is shown in the right graph; the number (n) of animals is for each genotype with MO. (D) Quantification of the SA-β-gal intensities shown in (B). Quantification of data presented in panel B (n = 12) is shown in the right graph; the number (n) of animals is for each morphant in genotype. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
Impact of Beclin 1 depletion on Spns1 deficiency in the presence or absence of p53. (A) Yolk opaque phenotype of control MO-injected or beclin 1 MO-injected wild-type (spns1+/+;tp53+/+), tp53zdf1/zdf1 (tp53m/m), spns1hi891/hi891 (spns1-/-), and spns1hi891/hi891;tp53zdf1/zdf1 (spns1-/-;tp53m/m) animals is compared at 48 hpf. Opacity is greater in the p53 mutant background with Spns1 deficiency. The attenuated suppressive effect of beclin 1 MO (12 ng/embryo) yolk opacity in spns1hi891/hi891;tp53zdf1/zdf1 animals is shown. Scale bar, 250 μm. (B) spns1hi891/hi891 animals coinjected with beclin 1 MO and p53 MO or beclin 1 MO and inverse-sequence p53 MO (inv. p53 MO; negative control) were assayed for the SA-β-gal detection at 84 hpf. The beclin 1 MO-mediated suppression of SA-β-gal in spns1hi891/hi891 animals was attenuated by p53 MO injection. Scale bar, 250 μm. (C) Quantification of the SAβ-gal intensities shown in (B). Quantification of data presented in panel B (n = 10) is shown in the right graph; the number (n) of animals is for each morphant. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
Impact of UV-induced apoptosis and autophagy on Spns1 deficiency in the presence or absence of p53. (A) UV-induced apoptosis can be detectable in either spns1+/+ or spns1hi891/hi891 animals in similar manners only under the normal p53 condition. The UV (18 mj/cm2) treatment was done at 60 hpf, followed by the phenotype observations in periderm or basal epidermal cells in the caudal eye at 72 hpf. Scale bar in image in top row, 250 µm. Scale bar in image in lower rows, 10 μm. (B) UV-induced autophagy enhances autolysosomal formation in spns1hi891/hi891 animals in the presence of p53. The UV (18 mj/cm2) treatment was done at 69 hpf, followed by the phenotype observations in periderm or basal epidermal cells in the caudal fin at 72 hpf. Scale bar, 10 µm. (C) Quantification of the EGFP-LC3 and LysoTracker fluorescence intensities shown in (B). Quantification of data presented in panel B (n = 6) is shown in the right graph; the number (n) of animals is for each genotype with MO. Three independent areas (periderm or basal epidermal cells in the caudal fin) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
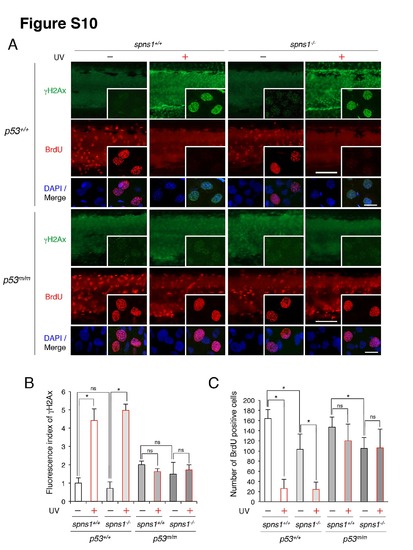
Detection of DNA damage response and DNA synthesis in spns1 mutants in the presence or absence of p53. (A) γH2AX- and BrdU detection in spns1 mutants in p53- and DNA damage-dependent manners. As shown in the green fluorescent panels, unaltered γH2AX intensities between spns1+/+ and spns1hi891/hi891 (spns1-/-) were apparent irrespective of p53 status without UV irradiation. Increased γH2AX intensities in response to UV irradiation were observed in the presence of p53 regardless of Spns1 status. Of note, certain basal increases of γH2AX intensities were detected in the p53 mutant background. As shown in the red fluorescent panels, reduced BrdU incorporation in spns1hi891/hi891 animals was detected in either normal or mutant p53 condition in the absence of UV treatment. UV-induced inhibition of DNA synthesis (reduction of BrdU signals) is apparently seen only in the normal p53 situation. The UV (18 mj/cm2) treatment was done at 68 hpf, followed by the phenotype observations at 72 hpf. Scale bar in the large image, 250 μm. Scale bar in the small merged image and inset, 10 μm. (B) Quantification of the γH2AX fluorescence intensities shown in (A). Quantification of data presented in panel A (n = 12) is shown in the right graph; the number (n) of animals is for each genotype. Three independent areas (periderm or basal epidermal cells in the trunk) were selected from individual animals. (C) Quantification of the BrdU-positive cells [in 25.6±2.2×104 μm areas; the trunk region starting from the rostral start point of the yolk extension (the distal end of the yolk) through the end of the caudal fin] shown in (A). Error bars represent the mean ± S.D., **p<0.005; *p<0.05; ns, not significant. |
|
Detection of mitotic cells in spns1 mutants in the presence or absence of p53. (A) Phosphorylated histone H3 (pH 3) staining in spns1-mutant animals with normal or mutant p53 backgrounds. The UV (18 mj/cm2) treatment was done at 68 hpf, followed by the phenotype observations at 72 hpf. Scale bar, 250 μm. (B) Quantification of the pH 3-positive cells [in 27.2±3.2×104 μm areas; the trunk region starting from the rostral start point of the yolk extension (the distal end of yolk) through the end of the caudal fin] shown in (A). Quantification of data presented in panel A (n = 9) is shown in the right graph; the number (n) of animals is for each genotype. Three independent areas (periderm or basal epidermal cells in the trunk) were selected from individual animals. Reduction of the pH 3 level was statistically significant in spns1hi891/hi891 (spns1-/-) animals in the presence of p53, and a reduced tendency (with no statistical significance) was also observed in spns1 mutants. Error bars represent the mean ± S.D., **p<0.05; *p<0.01; ns, not significant. |
|
Impact of UV irradiation on embryonic SA-β-gal activity in p53 and/or spns1 mutants. (A) Effect of UV treatment on embryonic SA-β-gal activity was validated in spns1-mutant animals with normal or mutant p53 backgrounds. The UV (18 mj/cm2) treatment was done at 68 hpf, followed by the phenotype observations at 72 hpf. Scale bar, 250 μm. (B) Quantification of the SA-β-gal intensities shown in (A). Quantification of data presented in panel A (n = 12) is shown in the right graph; the number (n) of animals is for each genotype. Error bars represent the mean ± S.D., **p<0.05; *p<0.01; ns, not significant. |
|
Suppression of spns1-mutant phenotypes by BafA treatment in zebrafish embryos. Suppression of yolk opacity by treatment with BafA (200 nM; 12 h treatment from 48 hpf through 60 hpf) in pigmented (AB line) and unpigmented (casper line) zebrafish embryos is shown. Scale bar, 250 μm. |
|
Suppression of spns1-mutant phenotypes by knockdown of the atp6v0c gene in zebrafish embryos. (A) Gross morphology, EGFP-LC3 and LysoTracker intensities in wild-type (wt) and spns1-mutant animals injected with atp6v0c MO (4 ng/embryo) at 48 hpf. Suppression of yolk opacity and SA-β-gal (SABG) by injection of atp6v0c MO in zebrafish embryos was observed at 48 and 60 hpf, respectively. Scale bar, 250 μm. (B) Quantification of the SA-β-gal intensities shown in (A). Quantification of data presented in panel A (n = 10) is shown in the right graph; the number (n) of animals is for each genotype with MO. (C) Effect of the PPIs (omeprazole; OPZ, lansoprazole; LPZ, and pantoprazole; PPZ) on embryonic senescence (SABG; SA-β-gal) in the spns1 mutant at 48 hpf. The drug treatments were done for 12 h from 36 hpf through 48 hpf. Scale bar, 250 μm. Error bars represent the mean ± S.D., *p<0.005; ns, not significant. |
|
Validations of lysosomal biogenesis and acidity in zebrafish embryos. (A) Whole-mount double staining with LysoTracker (10 μM, DND-99; red) and LysoSensor 189 (1 μM, DND-189; green). Live animals at 72 hpf were counterstained with LysoTracker and acidic pH-sensitive LysoSensor 189, simultaneously. LysoSensor 189 weakly detects acidic lysosomal signals in the spns1-mutant animals. Scale bar, 250 μm. Quantification of data presented for LysoSensor 189 (green) and LysoTracker (red) signals in panel A (n = 12) is shown in the right graph; the number (n) of animals is for each genotype. (B) Whole-mount double staining with LysoTracker (10 μM, DND-99; red) and LysoSensor 153 (1 μM, DND-153; green). Animals at 72 hpf were simultaneously counterstained by LysoTracker and neutral pH-sensitive LysoSensor 153. LysoSensor 153 can detect relatively neutral lysosomal signals in the spns1-mutant animals. Scale bar, 250 μm. Quantification of data presented for LysoSensor 153 (green) and LysoTracker (red) signals in panel B (n = 12) is shown in the right graph; the number (n) of animals is for each genotype. (C) Acidic pH-sensitive LysoSensor 189 (1 μM, green) probe in combination with LysoTracker (10 μM, red) was used in wt and spns1-mutant animals, and detectable signals in cells were obtained at 72 hpf. In wt fish treated with pepstatin A and E-64-d (P/E) (5 µg/ml each for 12 h), autolysosomal and/or lysosomal compartments were more prominently detected by LysoSensor 189 at the cellular level with enhanced accumulation of enlarged compartments under the identical LysoTracker staining condition. In contrast, in spns1-mutant animals, the cellular compartments were only weakly detectable by LysoSensor 189. Importantly, the short-term BafA treatment (for 1 h) largely attenuated or abolished staining of acidic compartments by both LysoSensor and LysoTracker, indicating that these autolysosomal and lysosomal compartments in wt animals treated with pepstatin A and E-64-d may retain some strong (lower pH) acidity. Scale bar, 10 μm. Quantification of data presented for LysoSensor 189 (green) and LysoTracker (red) signals in panel C (n = 12) is shown; the number (n) of animals is for each genotype with DMSO, pepstatin A and E-64-d (P/E) and/or BafA (+BafA; 1 h treatment). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (D) Using neutral pH-sensitive LysoSensor 153 (green) probes in combination with LysoTracker (red), wt and spns1-mutant animals were examined for detectable signals in cells when stained at 72 hpf. In spns1-mutant animals, autolysosomal and/or lysosomal compartments were more prominently detected by LysoSensor 153 at the cellular level with enhanced accumulation of enlarged compartments. In stark contrast, the cellular compartments in wt fish treated with pepstatin A and E-64-d (P/E) (5 μg/ml each for 12 h) were less detectable by LysoSensor 153 under the same staining condition used with LysoTracker. The short-term BafA treatment (for 1 h) still abolished the acidic compartments stained by both LysoSensor and LysoTracker, suggesting that the autolysosomal and lysosomal compartments observed in spns1-mutants may still retain some weak (higher pH) acidity. Scale bar, 10 µm. Quantification of data presented for LysoSensor 153 (green) and LysoTracker (red) signals in panel D (n = 12) is shown; the number (n) of animals is for each genotype with DMSO, pepstatin A and E-64-d (P/E) and/or BafA (+BafA; 1 h treatment). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. Error bars represent the mean ± S.D., *p<0.005; ns, not significant in (A), (B) and (D), and **p<0.005; *p<0.05; ns, not significant in (C). |
|
Validations of autolysosome formation and lysosomal biogenesis in zebrafish embryos. (A) Gross morphologies of BafA (100 nM)-treated or pepstatin A (5 μg/ml)- and E-64-d (5 μg/ml)-co-treated (P/E) wt [Tg(CMV:EGFP-LC3)] and spns1-mutant [Tg(CMV:EGFP-LC3); spns1hi891/hi891] animals. Embryos at 60 hpf were incubated with BafA or P/E for 12 h, and stained with LysoTracker at 72 hpf. Scale bar, 250 μm. Quantification of data presented in the middle and bottom rows (green; EGFP, red; mCherry) in panel A (n = 12) is shown; the number (n) of animals is for each genotype with DMSO, BafA or pepstatin A and E-64-d (P/E). (B) Intracellular autolysosome formation and lysosomal biogenesis in vehicle (DMSO)-treated, BafA (100 nM)-treated or pepstatin A (5 μg/ml)- and E-64-d (5 μg/ml)-treated (P/E) wt [Tg(CMV:EGFP-LC3)] and spns1-mutant [Tg(CMV:EGFP-LC3);spns1hi891/hi891] animals. Numerous large EGFP-LC3 puncta are evident in the BafA-treated embryos, with minimal LysoTracker staining. Some increased EGFP-LC3 speckles and strong enhancement of enlarged LysoTracker signals are evident in the cells from P/E-treated embryos. The same samples analyzed in (A) were observed by using confocal microscopy at a high magnification (×600). Scale bar, 10 μm. Quantification of data presented in the middle and bottom rows (green; EGFP, red; mCherry) in panel A (n = 6) is shown. The number (n) of animals is for each genotype with DMSO, BafA or pepstatin A and E-64-d (P/E). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. (C) Impaired autolysosomal acidification in BafA-treated wt or in spns1-mutant embryos, but not in pepstatin A- and E-64-d-treated (P/E) wt embryos. EGFP-LC3 and mCherry-LC3 double-transgenic wt [Tg(CMV:EGFP-LC3:mCherry-LC3)] and spns1-mutant [Tg(CMV:EGFP-LC3:mCherry-LC3);spns1hi891/hi891] zebrafish were used to monitor autolysosome formation. Embryos at 60 hpf were incubated with BafA (100 nM) or P/E (5 μg/ml each) for 12 h, to be observed later at 72 hpf. Quenching of EGFP-LC3 signals but not mCherry-LC3 signals is seen in the P/E-treated embryos, whereas unquenched EGFP-LC3 signals are evident in the BafA-treated as well as the spns1 MO-injected embryos. Whole-mount samples were observed by using confocal microscopy at a high magnification (×600). Scale bar, 10 μm. Quantification of data presented in the middle and bottom rows (green; EGFP, red; mCherry) in panel A (n = 6) is shown; the number (n) of animals is for each genotype with DMSO, BafA or pepstatin A and E-64-d (P/E). Three independent areas (periderm or basal epidermal cells above the eye) were selected from individual animals. |