- Title
-
CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- Authors
- Arjona, F.J., de Baaij, J.H., Schlingmann, K.P., Lameris, A.L., van Wijk, E., Flik, G., Regele, S., Korenke, G.C., Neophytou, B., Rust, S., Reintjes, N., Konrad, M., Bindels, R.J., Hoenderop, J.G.
- Source
- Full text @ PLoS Genet.
|
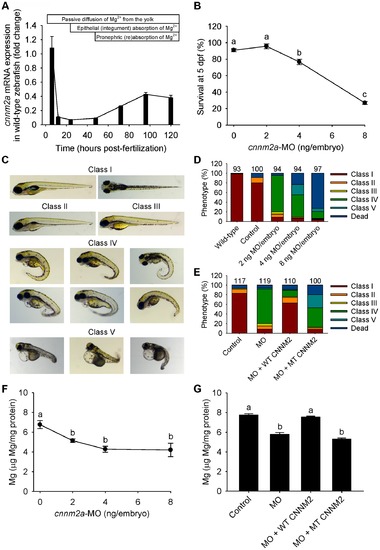
Knockdown of cnnm2a results in Mg wasting in zebrafish larvae (5 dpf). (A) mRNA expression of cnnm2a in developing zebrafish. Expression patterns were analysed by RT-qPCR (n = 6 per time point). (B) Survival curve at 5 dpf (n = 3 per experimental condition). The dose of zero represents injection with control-MO. (C) Morphological phenotypes in zebrafish larvae (5 dpf) in cnnm2a knockdown experiments. (D) Distribution of morphological phenotypes in zebrafish larvae (5 dpf) untreated (wild-type) or injected with different doses of cnnm2a-MO or control-MO. Numbers on top of the bars indicate the number of animals in each experimental condition. (E) Distribution of morphological phenotypes in zebrafish larvae at 5 dpf in rescue experiments. The wild-type phenotype (class I) was restored in morphants by co-injection of cnnm2a-MO (2 ng MO/embryo) with wild-type (WT) CNNM2 cRNA (50 pg cRNA/embryo), but not with mutant (MT, p.Glu357Lys) CNNM2 cRNA (50 pg cRNA/embryo). (F) Magnesium content in zebrafish injected with different doses of cnnm2a-MO, the dose of zero represents injection with control-MO (n = 10 per experimental condition except in 8 ng MO-injected zebrafish where n = 5). (G) Rescue of Mg wasting in morphant zebrafish by co-injection of cnnm2a-MO (2 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo). Co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo) did not restore Mg levels (n = 10 per experimental condition). Data are presented as mean ± SEM. Different letters indicate significant differences between mean values in experimental groups (P<0.05). |
|
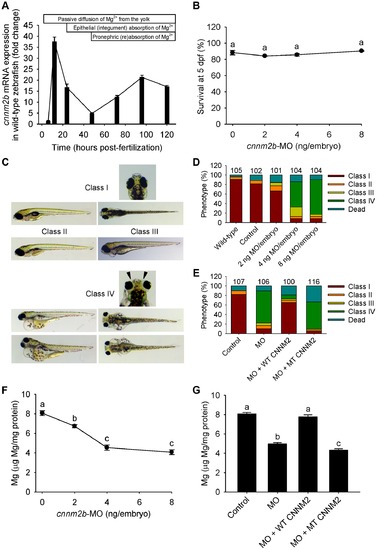
Knockdown of cnnm2b results in Mg wasting and brain malformations in zebrafish larvae (5 dpf). (A) mRNA expression of cnnm2b in developing zebrafish. Expression patterns were analysed by RT-qPCR (n = 6 per time point). (B) Survival curve at 5 dpf (n = 3 per experimental condition). The dose of zero represents injection with control-MO. (C) Morphological phenotypes in zebrafish larvae (5 dpf) in cnnm2b knockdown experiments. (D) Distribution of morphological phenotypes in zebrafish larvae (5 dpf) untreated (wild-type) or injected with different doses of cnnm2b-MO or control-MO. Brain malformations (widened cerebrospinal fluid spaces, class IV phenotype) are prominent in morphants injected with 4–8 ng MO/embryo. Numbers on top of the bars indicate the number of animals in each experimental condition. (E) Distribution of morphological phenotypes in zebrafish larvae at 5 dpf in rescue experiments. The wild-type phenotype (class I) was restored in morphants by co-injection of cnnm2b-MO (8 ng MO/embryo) with wild-type (WT) CNNM2 cRNA (50 pg cRNA/embryo), but not with mutant (MT, p.Glu357Lys) CNNM2 cRNA (50 pg cRNA/embryo). (F) Magnesium content in zebrafish injected with different doses of cnnm2b-MO. The dose of zero represents injection with control-MO (n = 10 per experimental condition). (G) Rescue of Mg wasting in morphant zebrafish by co-injection of cnnm2b-MO (8 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo). Co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo) did not restore Mg levels (n = 10 per experimental condition). Data are presented as mean ± SEM. Different letters indicate significant differences between mean values in experimental groups (P<0.05). |
|
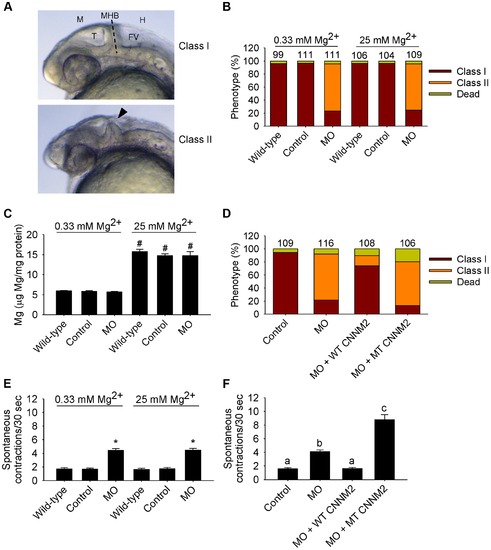
Dysfunctional cnnm2a causes brain abnormalities and increased spontaneous contractions in zebrafish embryos (25 hpf). (A) Phenotypes in zebrafish embryos untreated (wild-type) or following treatment with cnnm2a-MO (2 ng MO/embryo) or control-MO. Abbreviations indicate the following parts in the zebrafish embryonic brain: M, midbrain; T, tectum; MHB, midbrain-hindbrain boundary; FV, fourth ventricle; and H, hindbrain. (B) Distribution of phenotypes and (C) Mg content (n = 10 per experimental condition) in zebrafish embryos untreated (wild-type) or injected with 2 ng of cnnm2a-MO or control-MO and exposed to a medium with a concentration of Mg2+ of 0.33 or 25 mM. Numbers on top of the bars indicate the number of animals in each experimental condition. (D) Restoration of normal brain development by co-injection of cnnm2a-MO (2 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo), and not by co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo). (E) Spontaneous contractions in zebrafish embryos untreated (wild-type) or injected with 2 ng of cnnm2a-MO or control-MO and exposed to a medium with a concentration of Mg2+ of 0.33 or 25 mM (n = 30 per experimental condition). (F) Restoration of normal spontaneous contraction activity (n = 30 per experimental condition) by co-injection of cnnm2a-MO (2 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo), and not by co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo). Data are presented as mean ± SEM. *P<0.05 versus wild-type and control. #P<0.05 versus Mg2+-normal (0.33 mM Mg2+) medium. Data are presented as mean ± SEM. Different letters indicate significant differences between mean values in experimental groups (P<0.05). PHENOTYPE:
|
|
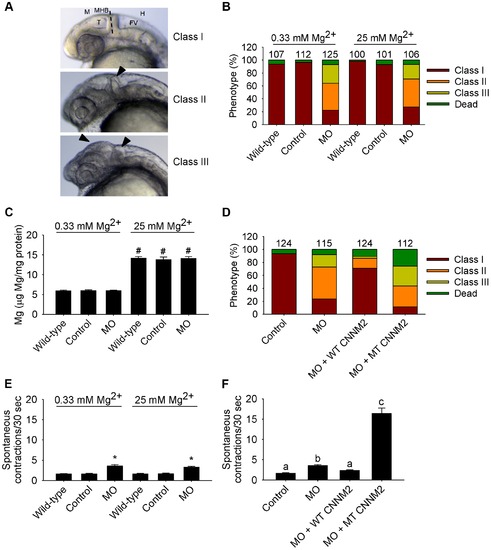
Dysfunctional cnnm2b causes brain abnormalities and increased spontaneous contractions in zebrafish embryos (25 hpf). (A) Phenotypes in zebrafish embryos untreated (wild-type) or following treatment with cnnm2b-MO (8 ng MO/embryo) or control-MO. See Figure 6 for an explanation of the abbreviations shown. (B) Distribution of phenotypes and (C) Mg content (n = 10 per experimental condition) in zebrafish embryos untreated (wild-type) or injected with 8 ng of cnnm2b-MO or control-MO and exposed to a medium with a concentration of Mg2+ of 0.33 or 25 mM. Numbers on top of the bars indicate the number of animals in each experimental condition. (D) Restoration of normal brain development by co-injection of cnnm2b-MO (8 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo), and not by co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo). (E) Spontaneous contractions in zebrafish embryos untreated (wild-type) or injected with 8 ng of cnnm2b-MO or control-MO and exposed to a medium with a concentration of Mg2+ of 0.33 or 25 mM (n = 30 per experimental condition). (F) Restoration of normal spontaneous contraction activity (n = 30 per experimental condition) by co-injection of cnnm2b-MO (8 ng MO/embryo) with cRNA encoding for wild-type (WT) CNNM2 (50 pg cRNA/embryo), and not by co-injection with cRNA encoding for mutant (MT, p.Glu357Lys) CNNM2 (50 pg cRNA/embryo). Data are presented as mean ± SEM. *P<0.05 versus wild-type and control. #P<0.05 versus Mg2+-normal (0.33 Mm Mg2+) medium. Data are presented as mean ± SEM. Different letters indicate significant differences between mean values in experimental groups (P<0.05). PHENOTYPE:
|
|
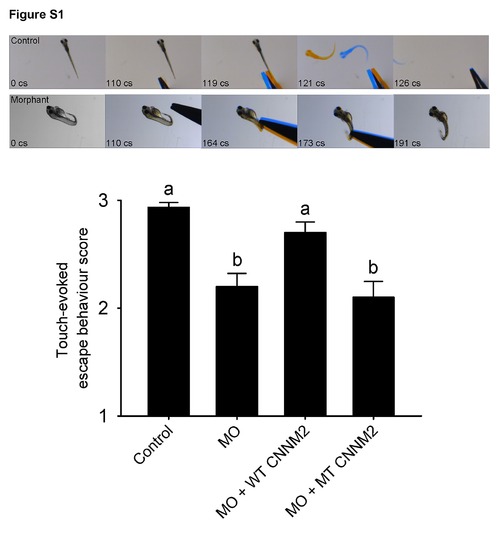
Impairment of touch-evoked escape behaviour in cnnm2a morphant zebrafish. Touch-evoked escape behaviour score in zebrafish cnnm2a morphants at 5 dpf after injection of 2 ng control-MO/embryo, 2 ng cnnm2a-MO/embryo, 2 ng cnnm2a-MO/embryo+50 pg wild-type (WT) CNNM2 cRNA/embryo, or 2 ng cnnm2a-MO/embryo+50 pg mutant (MT, p.Glu357Lys) CNNM2 cRNA/embryo. Three categories were distinguished, responders, late responders and non-responders, to which the following scores were given: 3 points for responders: fish quickly react (swimming or flicking the tail) to the stimuli after 1 or 2 twitches; 2 points for late responders: fish react (swimming or flicking the tail) to the stimuli after 3, 4 or 5 twitches; and 1 point for non-responders: fish do not react to the stimuli after more than 5 twitches. The upper part of the figure shows frames of videos showing touch-evoked escape contractions at 5 dpf of control and morphant zebrafish larvae. Time of each video frame is indicated in centisenconds (cs). Data are shown as mean ± SEM (n = 30). Different letters indicate significant differences between mean values in experimental groups (P<0.05). PHENOTYPE:
|
|
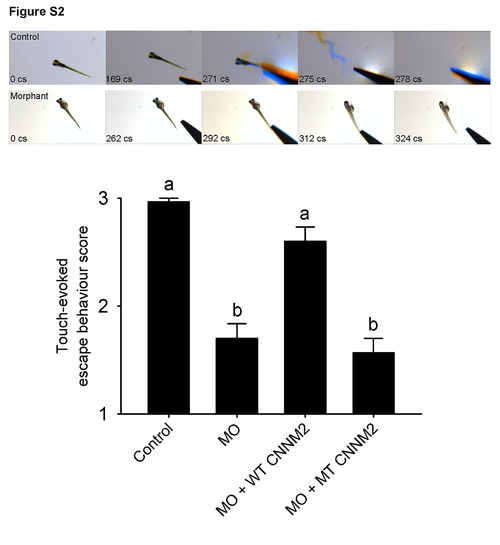
Impairment of touch-evoked escape behaviour in cnnm2b morphant zebrafish. Touch-evoked escape behaviour score in zebrafish cnnm2b morphants at 5 dpf after injection of 8 ng control-MO/embryo, 8 ng cnnm2b-MO/embryo, 8 ng cnnm2b-MO/embryo+50 pg wild-type (WT) CNNM2 cRNA/embryo, or 2 ng cnnm2b-MO/embryo+50 pg mutant (MT, p.Glu357Lys) CNNM2 cRNA/embryo. Three categories were distinguished, responders, late responders and non-responders, to which the following scores were given: 3 points for responders: fish quickly react (swimming or flicking the tail) to the stimuli after 1 or 2 twitches; 2 points for late responders: fish react (swimming or flicking the tail) to the stimuli after 3, 4 or 5 twitches; and 1 point for non-responders: fish do not react to the stimuli after more than 5 twitches. The upper part of the figure shows frames of videos showing touch-evoked escape contractions at 5 dpf of control and morphant zebrafish larvae. Time of each video frame is indicated in centisenconds (cs). Data are shown as mean ± SEM (n = 30). Different letters indicate significant differences between mean values in experimental groups (P<0.05). PHENOTYPE:
|