- Title
-
A novel conserved evx1 enhancer links spinal interneuron morphology and cis-regulation from fish to mammals
- Authors
- Suster, M.L., Kania, A., Liao, M., Asakawa, K., Charron, F., Kawakami, K., and Drapeau, P.
- Source
- Full text @ Dev. Biol.
|
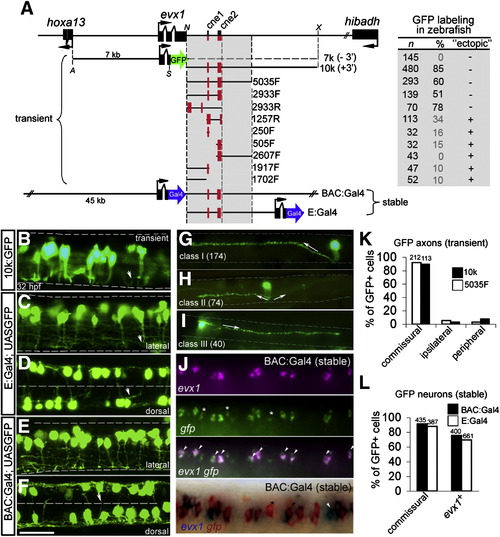
The pufferfish evx1 genomic region contains a downstream enhancer that labels evx1+ spinal commissural interneurons in zebrafish embryos. (A) Pufferfish evx1 locus and reporter constructs. Left, schematic of pufferfish evx1 reporter constructs. Black boxes represent exons. Translation start and orientation is indicated by arrows. For transient expression, farnesylated GFP and polyA (not shown) was fused in frame with exon 2 of evx1 (excluding homeodomain). 7 k contains the upstream promoter and lacks 3′ sequences (- 3′). 10 k contains a 10 kb downstream fragment (+ 3′) and others PCR fragments (size given in bp) downstream of GFP. Orientation is given as F (forward) or R (reverse). Restriction sites used were A (ApaI), N (NotI), S (SmaI) and X (XhoI). For stable transgenes, Gal4 was fused in frame with pufferfish evx1 using a ∼ 65 kb BAC and plasmid constructs (see Materials and methods). Right, observed frequencies of transient labeling in 28–32 hpf embryos. n, GFP+ embryos, and %, proportion with GFP+ interneurons. Frequent labeling of non-neuronal cells is noted under “ectopic”. (B–F) GFP-labeled interneurons (32 hpf) at rostral spinal cord levels with pufferfish evx1 constructs. B, C, E are lateral views and D, F are dorsal views. ∼ 2.5 segments are shown in each image. The limits of the spinal cord are indicated in B, C, E and the midline in D, F by dashed lines. Dorsal is up and rostral is to the left. Most neurons extend an axon (arrowhead in B, C) ventrally before crossing to the opposite side (out of focus). Scale bar: 35 μm. (G, H, I) Single-cell morphology of spinal interneurons labeled by 10k:GFP and 5035F:GFP constructs. (G) Class I (174 cells), commissural interneuron with large dorsal soma. Axon extends ventrally, crosses and then ascends dorsally 2–5 segments (arrow). (H) Class II (74 cells), commissural interneuron with medial soma whose axon extends ventrally before crossing and bifurcating (arrows). (I) Class III (40 cells), commissural interneuron with medial soma and a descending axon (arrow). (J) Dual ISH of evx1 (magenta) and gfp (green) mRNA in the spinal cord of stable transgenic BAC:Gal4; UAS:GFP embryos (fluorescent ISH, top panel and chromogenic ISH, bottom panel). Asterisks mark gfp+ cells that do not co-express evx1 and arrowheads mark cells labeled by evx1. In the bottom panel, a white arrowhead marks a cell that expresses only evx1. (K) Quantitation of axonal morphology of GFP+ spinal neurons labeled by 10k:GFP (10 k) and 5035F:GFP (5035F) in transient assays (shown in B). GFP+ cells were grouped as commissural, ipsilateral or peripheral based on the axonal projection. The total number of GFP+ cells is indicated above the bars. (L) Quantitation of axonal morphology and evx1 mRNA co-expression of spinal neurons labeled in BAC:Gal4; UAS:GFP and E:Gal4; UAS:GFP double transgenic embryos. Percentage (%) of GFP+ spinal neurons with commissural axons and gfp+ cells double labeled by evx1 mRNA are plotted. The total number of cells is indicated above the bars. EXPRESSION / LABELING:
|
|
Zebrafish and mouse Evx1 downstream fragments direct gene expression to chick V0 interneurons. (A) Schematic of the Evx1 genomic region and the fragments used to generate the constructs for electroporation in the chick. A 3.4 kb zebrafish fragment including the last 89 bp of exon 3, 3′ UTR and CNE1 and CNE2 was placed upstream of the tkGFP cassette. The mouse reporter construct consists of 3.6 kb upstream and 4.6 kb downstream. YFP was fused in frame to exon 1 (+ 156 from ATG start) in the latter. St, StuI, B, BamHI, Na, NaeI and A, ApaI. (B–G) Representative sections from electroporated embryos. Pufferfish (B), zebrafish (C) and mouse (D) constructs show a highly similar pattern of GFP or YFP expression localized to a cluster of commissural interneurons (arrowheads) whose axons can be seen crossing to the opposite side of the cord (arrows). The outline of the spinal cord is drawn as dashed lines. Scale bar: 110 μm. (E,F,G) Magnified views of GFP+ cells double labeled with Evx1/Evx2 antibodies. Medial (m) and lateral (l) orientation is indicated by the double arrow and is the same for F and G. Percentage of GFP+ cells that co-expressed Evx1/Evx2 on average is indicated at the bottom left corner of each panel (see text for details). At least 6 embryos were examined for each construct. |
|
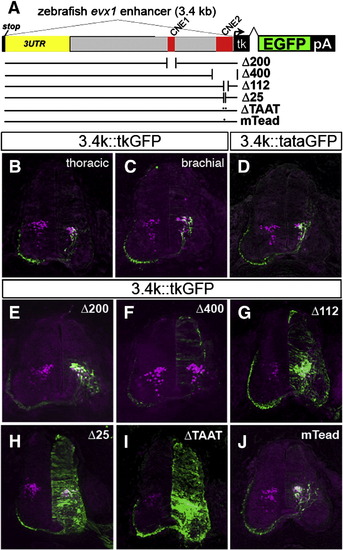
Deletion analysis of the zebrafish evx1 enhancer in the chick neural tube. (A) Schematic of deletions and mutations in the zebrafish 3.4 kb (3.4 k) downstream fragment driving GFP with a minimal tk promoter in the chick. Deletions Δ200 (NheI/PacI, deletes CNE1), Δ400 (KpnI, deletes CNE2), were generated by digestion and religation. Δ112 (deletes CNE2), Δ25 (deletes 25HCS) and ΔTAAT (deletes HD in 25HCS) were made by PCR deletion mutagenesis (Table S2). In mTead, a putative Tead2 site was mutated (TGAATG to ATTCCG). Sections of electroporated embryos were double labeled with antibodies to GFP and Evx1/Evx2. (B,C) Control sections at thoracic and brachial spinal cord levels. The outline of the spinal cord is drawn as dashed lines. Scale bar: 100 μm. (D) 3.4 kb::tataGFP (with TATA promoter instead) generates an almost identical pattern as with control tk reporter. (E) Δ200 retains V0 labeling and similar specificity to controls. (F) Δ400 eliminates V0-specific labeling but shows low-level background expression. (G) Δ112 retains V0 labeling but shows also widespread expression in the neural tube. (H, I) Δ25 and ΔTAAT also generate widespread GFP expression. (J) Mutation of the putative Tead2 (mTead) site does not alter the pattern of GFP expression. Sections are representative from analysis of 5–9 embryos from each construct. |
|
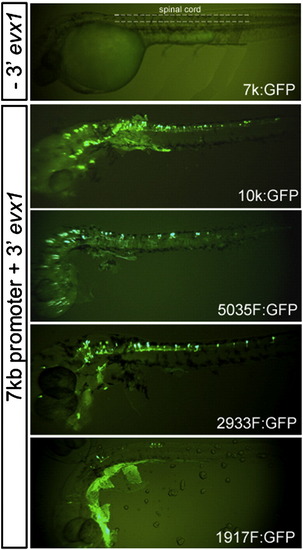
Transient expression of pufferfish evx1:GFP reporter constructs in zebrafish embryos. All reporter constructs contained a 7 kb promoter and GFP fused in frame in exon 2 as described in Fig. 2A. Constructs with or without the 3′ evx1 region (- 3′ evx1 and + 3′ evx1) were injected into the cytoplasm of one-cell stage embryos. Fluorescence images of whole-mount living embryos were taken under a dissection microscope at ∼ 32 hpf. The region of the ‘spinal cord’ examined in our study is labeled in A and the dorso-ventral limits of the spinal cord marked by dash lines. Constructs 7 k (7 kb promoter alone), 10k, 5035F, 2933F and 1917F are described in Fig. 2A. Dorsal is up and rostral is to the left. |
|
Deletions in the zebrafish evx1 enhancer cause de-regulation of reporter GFP expression in the chick. Embryos were electroporated with deletion variants of 3.4 kb::tkGFP (from Fig. 4A). Δ200 removes CNE1, Δ400 removes CNE2, Δ25 removes 25HCS and ΔTAAT is a double deletion of two homeodomain (TAAT) sites in 25HCS. (B) Low magnification image of GFP in a control (3.4 kb) whole-mount chick embryo. Bright GFP signal is observed along the rostral (r) to caudal (c) axis of the trunk (double arrow in B,C) within the spinal cord. (C–G) High magnification images focusing on a side view of the spinal cord. (C) control 3.4 kb. (D) Δ200 is indistinguishable from the control. (E) Δ400 dramatically eliminated GFP expression. (F, G) Δ25 and ΔTAAT ectopically labeled peripheral motor axon bundles that exit the spinal cord (arrowheads). |
Reprinted from Developmental Biology, 325(2), Suster, M.L., Kania, A., Liao, M., Asakawa, K., Charron, F., Kawakami, K., and Drapeau, P., A novel conserved evx1 enhancer links spinal interneuron morphology and cis-regulation from fish to mammals, 422-433, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.