- Title
-
Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish
- Authors
- Lin, C.Y., Yung, R.F., Lee, H.C., Chen, W.T., Chen, Y.H., and Tsai, H.J.
- Source
- Full text @ Dev. Biol.
|
Tagging all head muscles by using the transgenic zebrafish line Tg(α-actin:RFP). Endogenous α-actin transcripts (A) and red fluorescent protein (RFP) reporter transcripts (B) were detected by whole-mount in situ hybridization of zebrafish embryos at 14 hpf (lateral view: A, B). RFP expressions in the embryos derived from the transgenic line (α-actin:RFP) at 22.5 hpf (C, D), 72 hpf (E, F), and 120 hpf (G, H) were observed from a lateral view (E, G) and from a ventral view (F, H). Panel D is a magnification of panel C. RFP appeared in the formed somites (D, arrowhead) but was absent in the newly forming somite (D, arrow) at 22.5 hpf. Meanwhile, all cranial muscles were labeled by red fluorescent signal in the embryos both at 72 and at 120 hpf. The muscles are designated following the scheme of Schilling and Kimmel (1997): ah, adductor hyoideus; am, adductor mandibulae; ao, adductor operculi; do, dilator operculi; dpw1–5, dorsal pharyngeal wall 1–5; hh, hyohyoideus; ih, interhyoideus; ima, intermandibularis anterior; imp, intermandibularis posterior; io, inferior oblique; ir, inferior rectus; lap, levator arcus palatini; lr, lateral rectus; mr, medial rectus; sh, sternohyoideus; so, superior oblique; sr, superior rectus; and tv 1–5, transvs. ventralis 1–5. EXPRESSION / LABELING:
|
|
The temporal expressions of myf5 and myod during cranial muscle development. The temporal expressions of myf5 (A–C, E, G, I) and myod (D, F, H, J) transcripts were analyzed by whole-mount in situ hybridization in zebrafish embryos. The transcript of myf5 was detected in the craniofacial region at 24 hpf (A, arrow); in the io, the precursor of lr, and 1st arch (lr/1st arch) at 30 hpf (B); in the so, io, lr, 1st, 2nd, and 3rd arches at 32 hpf (C); in the so, io, sh, 2nd, and 3rd arches at 36 hpf (E); and in the so, io, and sh at 42–48 hpf (G, I). myf5 was not expressed in the lr and 1st arch at 36 hpf (E). Meanwhile, myod was expressed in the craniofacial muscles mr/ir/sr, lr, and 1st arch at 32 hpf (D); in the mr/ir, sr, lr, MP, IM, CHD, and CHV at 36 hpf (F); and in all the cranial muscle at 42–48 hpf (H, J). CD: the constrictor dorsalis, which differentiates to lap and do; CHD: the constrictor hyoideus dorsalis, which differentiates to ah and ao; CHV: the constrictor hyoideus ventralis, which differentiates to ih and hh; IM: the intermandibularis, which differentiates to ima and imp; MP: the masticatory plate, which differentiates to CD and am. lr, lateral rectus; for other abbreviations, see the legend of Fig. 1. |
|
Myf5 is required for cranial muscle development, except the primordia of mr/ir/sr. Embryos derived from the transgenic line Tg(α-actin:RFP; A, B) and from the wild-type strain (C–J) were used. All the embryos were lateral views except panels E and F, which were dorsal–lateral view. The embryos injected with 4 ng of myf5-morpholino oligonucleotide (MO) to inhibit myf5 translation specifically, were studied to observe the development of cranial muscle (B) and the expression of myod, et1, and myogenin (D, F, H, J) at the stages indicated. Red fluorescent protein (RFP) signal was detected only in the mr/ir and sr primordia in the myf5 transgenic morphant (A vs. B). myod was expressed only in the mr/ir/sr primordia in the myf5 wild-type morphant at 36 hpf (C vs. D) and at 58 hpf (G vs. H). Similarly, myogenin was detected only in the mr/ir/sr primordia in myf5 morphant (I vs. J). However, the et1 transcript was changed little in the ventral arch mesoderm core in the myf5 morphant (E vs. F, arrows), indicating that the development of the ventral arch mesoderm core in myf5 morphant was normal. |
|
Myod is required for the development of posterior extraocular recti and ventral branchial muscles. Embryos derived from the transgenic line Tg(α-actin:RFP) (A–D) and from the wild-type strain (E–J) were used. Embryos injected with 4 ng of myod-morpholino oligonucleotide (MO) to inhibit specifically myod translation were followed to observe the development of cranial muscle (B, D) and the expression of myf5 (F, H) and myogenin (J; lateral views: A, B, E–J; ventral views: C, D) at the stages indicated. The anterior extraocular recti (io and so), dorsal branchial muscle (ah, ao, do, and lap), and sh developed normally in the myod transgenic morphant (A vs. B; C vs. D). The posterior extraocular (sr, mr, ir, and lr) and the ventral branchial muscle (ima, imp, ih, and hh) were totally lost. When wild-type embryos were injected with myod-MO, myf5 was expressed normally in the myod morphants at 30 hpf (E vs. F) and at 36 hpf (G vs. H). Whereas, the expression of myogenin was decreased in myod morphants at 58 hpf (I vs. J), indicating a reduction in myogenin-positive muscle fibers. |
|
Myf5 and Myod function independently to activate progenitor lineages in muscles of the head region. Embryos co-injected with 4 ng of myf5-morpholino oligonucleotide (MO) with 4 ng of myod-MO to inhibit specifically both myf5 and myod translation, respectively, were used to observe the development of cranial muscle (A, B) and the expression of myogenin and mrf4 (D, F) at 72 hpf. Panel B was magnified from the head area of panel A. No red fluorescent protein (RFP) signal was detected in muscle primordia in the myf5 and myod double-knockdown morphants derived from the transgenic line Tg(α-actin:RFP; A, B). Similarly, the transcripts of myogenin (C vs. D) and mrf4 (E vs. F) were not detected in the myf5 and myod double-knockdown morphants (I vs. J). The myod mRNA did not rescue the formation of RFP-labeled primordia muscles in myf5 morphants (G). Similarly, the myf5 mRNA did not rescue the formation of RFP-labeled primordia muscle in myod morphants (H). For abbreviations, see the legend of Fig. 1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of Myod up-regulated Myf5 expression, and vice versa, in the cranial muscle that migrated from the anterior somites. The expression of myf5 was increased in sternohyoideus (sh) primordia, fin bud (fb), and posterior hypaxial muscle (phm) that migrated from the anterior somites, when embryos received myod-morpholino oligonucleotide (MO; vs. B). This consequence was consistent in trunk myogenesis. There was weak myf5 expression in the trunk of wild-type embryos at 48 hpf (C). However, myf5 appeared predominantly in the dorsal (ds) and ventral (vs) parts of somites in the myod morphants (E), indicating that myodknockdown up-regulated the expression of myf5 both in sh, which originated from trunk somite, and in trunk muscle. Similarly, the expression of myod was also enhanced in the trunk of myf5 morphants, compared to wild-type embryos (D vs. F). The myogenin expression was similar to that of myf5 in myod morphants (E, G) and to that of myod in myf5 morphants (F, H). EXPRESSION / LABELING:
|
|
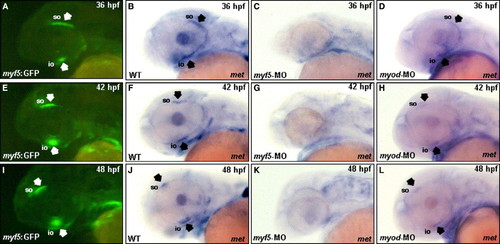
Myf5 is required for the forward migration of the superior oblique (so) and inferior oblique (io) primordia toward the anterior eye region. Embryos derived from the transgenic line Tg(myf5:EGFP), in which an upstream region of zebrafish myf5 was fused with enhanced green fluorescent protein (EGFP), were used to trace the migration of cranial muscle that express myf5. The primordia of io and so labeled with EGFP were clearly visible in the embryos derived from the transgenic line Tg(myf5:EGFP) at 36–48 hpf under fluorescent microscopy (A, E, I; arrows). Whole-mount in situ hybridization of wild-type zebrafish embryos showed that the expression of met, a cell marker of migration, was positive in the io and so at 36–48 hpf (B, F, J; arrows). Embryos injected with myf5-morpholino oligonucleotide (MO) and myod-MO to inhibit myf5 and myod translations, respectively, expressed met at the stages indicated. The met transcript was not expressed in the io and so of myf5 morphants (C, G, K), but met was expressed normally in the io and so of myod morphants (D, H, L; arrows). For abbreviations, see the legend of Fig. 1. |
|
Myf5 is required for the migration of the sternohyoideus (sh) primordia from anterior somites. The transcripts of myf5 (A), myod (B), and met (C) were detected in sternohyoideus (sh) premordia, fin bud (fb), and posterior hypaxial muscle (phm), that were derived from anterior somites at 36 hpf. Embryos injected with myf5-morpholino oligonucleotide (MO) and myod-MO to inhibit specifically myf5 and myod translations, respectively, expressed met at the stages indicated. The expression of met was almost lost in myf5 morphants; only slight signals were detected in the phm (D; arrow). However, met was expressed normally in myod morphants (E). At 48 hpf, myogenin was expressed in the sh, fb, and phm of wild-type (F) and myod morphants (H), but myogenin was not expressed in the sh, fb, or phm of myf5 morphants (G). Instead, the expression of myogenin was up-regulated in the trunk of myf5 morphants. Embryos derived from the transgenic line Tg(α-actin:RFP; I–K) were used. At 72 hpf, the sh, fb, and phm showing red fluorescent signal were clearly observed both in wild-type (I; arrow) and myod morphants (K; arrow) but not in myf5 morphants (J). For abbreviations, see the legend to Fig. 1. |
|
A plausible model to present the distinct modulation of Myf5 and Myod during craniofacial myogenesis of zebrafish. (A) Schematic illustration of the dynamic expression of myf5 and myod in the cranial muscle of zebrafish embryos at 32 and 42 hpf. The myf5- or myod-positive muscle fibers are labeled in red; myf5- and myod-negative ones are labeled in gray. (B) Schematic illustration of the presence (red) or absence (gray) of cranial muscle fibers in the embryos injected with either myf5-morpholino oligonucleotide (MO; left) or myod-MO (right) at 72 hpf. (C) Schematic diagram of all cranial muscles that are categorized into three groups (represented in red, blue, and green) on the basis of three regulatory pathways of myf5 and myod during development: (I) for so, io, sh, and dorsal arch, not only does myf5 modulate myogenesis directly to generate myogenesis at the basal level but myf5 also triggers myod expression to enhance myogenesis at a high level; (II) for lr and ventral arch, myf5 defines the cell fate of muscle and myod is the major factor of myogenesis; and (III) for sr, mr, and ir, myod modulates myogenesis directly. |
|
Spatiotemporal expression of myf5, myod, myogenin, α-actin and RFP during cranial muscle development. Embryos derived from the wild-type (A–T) and from the transgenic line Tg(α-actin:RFP) (U–Y) were observed by lateral view except that E, J, O, T, and Y were observed by dorsal view. Comparison the expression patterns of myf5 (A–E), myod (F–J), myogenin (K–O) and α-actin (P–T), we found that the myf5 transcripts in the superior oblique (so) and inferior oblique (io) were gradually decreased during 36 to 48 hpf. The myf5 transcripts were not detected in the head after 58 hpf (A–E). myod was expressed earlier than myogenin and α-actin in the cranial primordial muscle cells (F vs. K and P). The myod transcripts were present in most cranial muscles except io and so at 36 hpf; thereafter, myod was expressed all cranial muscles at 482Fi> in the cranial primordial muscle cells (F vs. K and P). The myod transcripts were present in most cranial muscles except hpf (G). myogenein was slightly expressed in the intermandibularis, masticatory plate, and io at 36 hpf (K), and were detected in all the cranial muscles similar to myod staining at 48 hpf (G vs. L). Whereas α-actin transcripts did not detectable until 58 hpf (V–W). The RFP expression in the Tg(α-actin:RFP) embryos was detectable at 55 hpf, and fluorescent signal became very prominent at 58 hpf EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 299(2), Lin, C.Y., Yung, R.F., Lee, H.C., Chen, W.T., Chen, Y.H., and Tsai, H.J., Myogenic regulatory factors Myf5 and Myod function distinctly during craniofacial myogenesis of zebrafish, 594-608, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.