Figure Caption
Fig. 3.
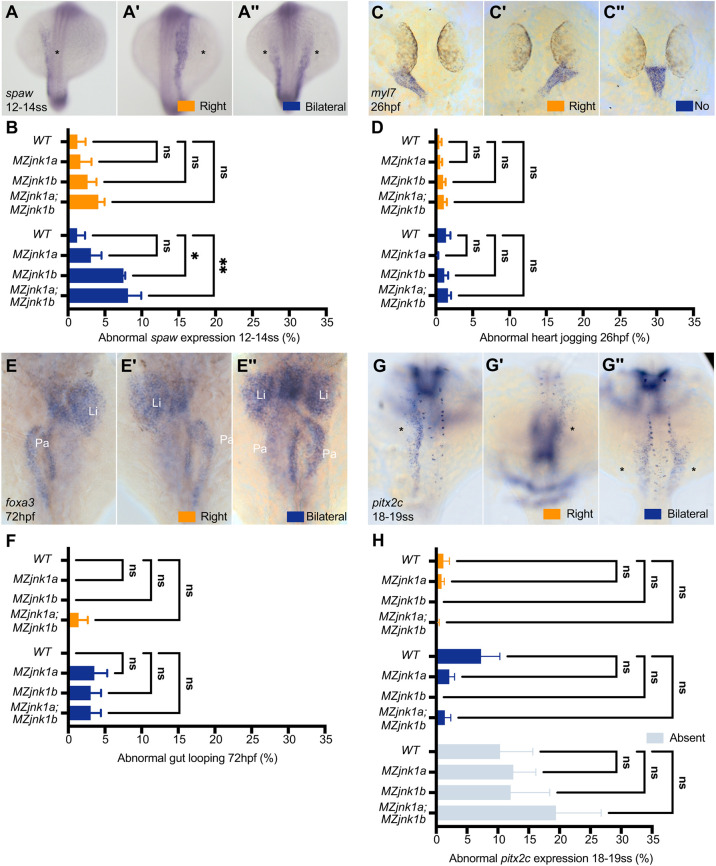
Loss of jnk1 disrupts Nodal signalling, but not organ asymmetry. (A-A″) Representative images of mRNA in situ hybridisation for the zebrafish Nodal homolog southpaw (spaw) at 12-14 ss, showing normal expression in the left lateral plate mesoderm (LPM) (A, asterisk), and abnormal right-sided (A′, asterisk) or bilateral (A″, asterisks) expression. (B) Characterisation of abnormal spaw expression in wild type, and in MZjnk1a, MZjnk1b and MZjnk1a;MZjnk1b mutants at 12-14 ss. MZjnk1b and MZjnk1a;MZjnk1b mutants display a significant increase in the percentage of embryos with bilateral spaw expression. Loss of jnk1a alone does not have a significant impact on spaw expression. (C-C″) Representative images of mRNA in situ hybridisation of the pan-cardiac marker myosin, light chain 7, regulatory (myl7) at 26 hpf showing normal left jogging of the heart (C), abnormal right jogging (C′) or no jogging (C″). (D) Quantification of jogging in wild type, and MZjnk1a, MZjnk1b and MZjnk1a;MZjnk1b mutants at 26 hpf. Loss of jnk1 does not affect heart jogging. (E-E″) Representative images of mRNA in situ hybridisation of the endodermal marker forkhead box A3 (foxa3) at 72 hpf showing organ placement of the liver (Li) and pancreas (Pa) following gut looping (E), reversed gut looping (E′) or a failure of LPM migration, resulting in a bilateral gut, most obviously observed by the presence of bilaterally positioned livers (E″). (F) Quantification of gut looping in wild type, and MZjnk1a, MZjnk1b and MZjnk1a;MZjnk1b mutants at 72 hpf. Loss of jnk1 does not affect endoderm morphogenesis. (G-G″) Representative images of mRNA in situ hybridisation of paired-liked homeodomain 2, isoform c (pitx2c) at 18-19 ss, showing normal expression in the left lateral plate mesoderm (F, asterisk), and abnormal right-sided (F′, asterisk) or bilateral (F″, asterisks) expression. (H) Characterisation of abnormal pitx2c expression in wild type, and MZjnk1a, MZjnk1b and MZjnk1a;MZjnk1b mutants at 18-19 ss. Loss of jnk1 does not result in abnormal expression of pitx2c. (B) Data are mean±s.e.m., two-way ANOVA comparison of right and bilateral. n=3 clutches. Minimum clutch sizes: wild type, n=27; MZjnk1a, n=21; MZjnk1b, n=27; MZjnk1a;MZjnk1b, n=17. (D) Data are mean±s.e.m., two-way ANOVA comparison of right jogging and no jogging. n=7 clutches for wild type, n=6 clutches for MZjnk1a, and n=8 clutches for MZjnk1b and MZjnk1a;MZjnk1b. Minimum clutch sizes: wild type, n=32; MZjnk1a, n=46; MZjnk1b, n=46; MZjnk1a;MZjnk1b, n=43. (F) Data are mean±s.e.m., two-way ANOVA comparison of right and bilateral. n=3 clutches. Minimum clutch sizes: wild type, n=25, MZjnk1a, n=16; MZjnk1b, n=21; MZjnk1a;MZjnk1b, n=22. (H) Data are mean±s.e.m., two-way ANOVA, multiple comparisons analysing right, bilateral and absent pitx2c expression. n=5 clutches. Minimum clutch sizes: wild type, n=75; MZjnk1a, n=73; MZjnk1b, n=64; MZjnk1a:MZjnk1b, n=37. (A-A″,C-C″,G-G″) Dorsal views. (E-E″) Ventral views. ns, not significant. *P<0.05, **P<0.01.
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development